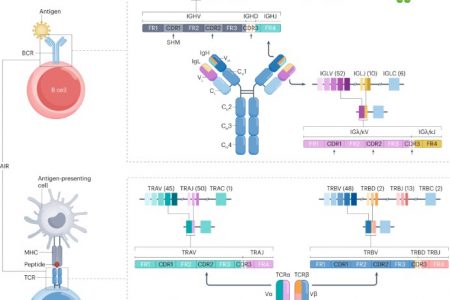
In immunology research, understanding the immune repertoire is crucial for developing better therapies, vaccines, and diagnostics. Two key components of the immune system, B-cell receptors (BCRs) and T-cell receptors (TCRs), play distinct yet interconnected roles in recognizing pathogens. Sequencing the BCR and TCR repertoires provides deep insights into immune responses. But what exactly differentiates these two receptors?
Let’s dive into the structural, functional, and Diversity differences between BCR and TCR.
Understanding BCRs and TCRs in Adaptive Immunity
The adaptive immune system is our body’s defense mechanism against pathogens. BCRs and TCRs are essential for this system, as they recognize and respond to foreign antigens. While both BCRs and TCRs serve as antigen receptors, they differ significantly in structure and function.
- BCRs are located on the surface of B-cells and primarily recognize intact antigens, such as proteins, polysaccharides, and lipids.
- TCRs, on the other hand, are found on T-cells and recognize peptide fragments presented by MHC molecules on the surface of other cells.
Understanding these differences is key to understanding immune system diversity and how the immune system adapts to new threats.
Structural Differences Between BCRs and TCRs
BCRs and TCRs have different structures that determine their roles in the immune system, as evidenced by recent structural studies and molecular analyses.BCRs are immunoglobulins (Ig), composed of two heavy and two light chains that form a Y-shaped molecule. The variable regions of these chains bind directly to the antigen. Recent cryo-EM studies have revealed the detailed structure of full-length transmembrane BCRs in their resting state. These studies show that the BCR complex is characterized by a 1:1 stoichiometry of two membrane-bound immunoglobulin (mIg) chains associated with Igα/Igβ subunits. The membrane-spanning pair of mIg chains adopt a Y-shaped topology, with flexible Fab fragments and Fc domains that tightly pack with the extracellular domains of the Igα/Igβ heterodimer.
TCRs consist of alpha and beta chains (or gamma and delta chains in some T-cells). These chains are linked by a disulfide bond and work together to bind to peptide-MHC complexes on the surface of antigen-presenting cells. Unlike BCRs, TCRs associate with three signaling dimers (CD3γε, CD3δε, ζ-ζ), forming a more complex transmembrane structure. The TCR-CD3 complex forms an eight-helix bundle in which the transmembrane helices of the three signaling dimers closely associate with the two TCR transmembrane helices.
Both BCRs and TCRs belong to the immunoglobulin-like fold family, with their antigen-binding sites composed of three loops called complementarity-determining regions (CDRs) in each receptor chain. The third CDR (CDR3) is the most diverse among the three CDRs due to V(D)J recombination and random nucleotide insertions during this process.
These structural differences between BCRs and TCRs reflect their distinct functions in the immune system. BCRs can recognize and bind to intact antigens directly, while TCRs recognize peptide fragments presented by MHC molecules on the surface of antigen-presenting cells.

Figure 1. Structure and activation of the BCR and TCR complexes. (Yufang Shi et al,. 2024)
Functional Differences: Antigen Recognition and Binding
The primary functional difference between BCRs and TCRs lies in the type of antigens they recognize, as demonstrated by numerous scientific studies.BCRs recognize whole, unprocessed antigens. They can bind to a wide variety of pathogen-associated molecular patterns (PAMPs), such as proteins or lipids, in their natural state. A study by Kovaltsuk et al. (2020) used computational methods to analyze the observed antibody space, demonstrating the diverse range of epitopes that BCRs can recognize, including both linear and discontinuous epitopes on intact antigens
1.TCRs recognize processed peptide fragments that are displayed by MHC molecules on the surface of cells. TCRs do not bind to free-floating antigens but rather to those that have been "processed" and presented by antigen-presenting cells. This was clearly demonstrated in a comprehensive review by Rossjohn et al. (2015), which detailed the structural basis of TCR recognition of peptide-MHC complexes.
2. This distinction is vital because it shapes how the immune system responds to pathogens and distinguishes humoral immunity (BCR-mediated) from cell-mediated immunity (TCR-mediated). A study by Newell and Davis (2014) highlighted how this difference in antigen recognition contributes to the distinct roles of B cells and T cells in the immune response.
3.The functional differences between BCRs and TCRs are further exemplified in their interactions with antigens. BCRs can directly interact with epitopes on intact antigens, as shown by Sela-Culang et al. (2013) in their analysis of antibody-antigen recognition.
4. In contrast, TCRs interact with a complex formed by the antigen-derived peptide and the MHC molecule, a process elucidated in detail by Garcia et al. (2012) in their structural studies of TCR-pMHC interactions.
5.These differences in antigen recognition and binding mechanisms between BCRs and TCRs are fundamental to the diverse and complementary roles of B cells and T cells in adaptive immunity, allowing for a comprehensive immune response to a wide range of pathogens.
Diversity Generation: V(D)J Recombination and Somatic Hypermutation
The diversity of both BCRs and TCRs is critical for recognizing a vast array of antigens. However, the mechanisms behind this diversity differ:BCRs undergo somatic hypermutation (SHM), a process that introduces mutations in the variable region of the immunoglobulin genes after activation, leading to an even greater variety of antigen-binding sites. This increases BCR diversity substantially. A study by Kovaltsuk et al. demonstrated the extensive diversity generated by SHM in BCRs, showing that it contributes significantly to the observed antibody space.
1. TCRs, on the other hand, rely only on V(D)J recombination to generate diversity. This recombination process rearranges gene segments (Variable, Diversity, and Joining segments) to form the unique TCRs. Research by Murugan et al. quantified the diversity generated by V(D)J recombination in TCRs, estimating a potential repertoire size of 10^15 to 10^20 unique receptors
2. While BCRs have higher diversity due to SHM, TCR diversity is mainly limited to the V(D)J recombination process, which, although highly diverse, doesn’t reach the same level as BCRs. A comparative study by Elhanati et al. highlighted this difference, showing that BCR repertoires exhibit higher mutation rates and greater diversity compared to TCR repertoires
3. The impact of these different diversification mechanisms is evident in the functional capabilities of B and T cells. BCRs can undergo affinity maturation through SHM, allowing for the production of high-affinity antibodies. This process was elucidated in detail by Victora and Nussenzweig, who described how SHM enables the selection of B cells with improved antigen binding
4. In contrast, TCRs maintain a relatively stable repertoire after initial generation. This stability was demonstrated by Qi et al., who showed that the TCR repertoire remains largely consistent over time, with diversity primarily generated during T cell development.

Figure 2: BCR and TCR Repertoire Diversity (Vanessa Mhanna et al,. 2024)
Summary and Conclusion
In summary, BCRs and TCRs are fundamental components of the adaptive immune system, each playing distinct roles in pathogen recognition and immune response. BCRs primarily recognize intact antigens in their natural form, while TCRs specifically bind to peptide fragments presented by MHC molecules. This difference in antigen recognition is reflected in their structural variations; BCRs are composed of two heavy and two light chains, forming a Y-shaped immunoglobulin, whereas TCRs consist of alpha and beta chains (or gamma and delta chains) that associate with signaling dimers.The mechanisms generating diversity in these receptors also differ significantly. BCRs undergo somatic hypermutation (SHM) after activation, enhancing their ability to bind a wide range of antigens through affinity maturation. In contrast, TCR diversity is generated solely through V(D)J recombination, resulting in a more stable repertoire post-development.Furthermore, the methodologies for sequencing these receptors vary, with BCR sequencing often requiring higher depths due to the greater diversity generated by SHM. Advanced computational tools play a crucial role in analyzing the complex data generated from both BCR and TCR sequencing, enabling researchers to uncover patterns related to antigen specificity and immune responses.Understanding these differences is essential for developing targeted therapies, vaccines, and diagnostics that leverage the unique properties of BCRs and TCRs. As immunology research continues to evolve, insights gained from studying these receptors will undoubtedly contribute to more effective strategies for combating diseases and enhancing immune responses.
To further explore the methodologies and applications in this field, the following topics provide a comprehensive guide:
Overview of Strategies for TCR profiling
TCR Detection Application and Method
Immune Repertoire Sequencing: Introduction, Workflow, and Applications
What is B-Cell Receptor Repertoire Sequencing?
T-Cell Receptor Sequencing Using RNA-Seq Protocol
Bulk Immune Repertoire (TCR/BCR) Sequencing: Principles and Applications
Cell Receptor Sequencing (TCR) Overview
References
- Wang Y, Mashock M, Tong Z, Mu X, Chen H, Zhou X, Zhang H, Zhao G, Liu B, Li X. Changing Technologies of RNA Sequencing and Their Applications in Clinical Oncology. Front Oncol. 2020 Apr 9;10:447. doi: 10.3389/fonc.2020.00447.
- Hong, M., Tao, S., Zhang, L. et al. RNA sequencing: new technologies and applications in cancer research. J Hematol Oncol 13, 166 (2020). https://doi.org/10.1186/s13045-020-01005-x
- Jiang, L., Chen, H., Pinello, L., & Yuan, G.-C. (2016). GiniClust: Detecting rare cell types from single-cell gene expression data with Gini index. Genome Biology, 17, Article 144. https://doi.org/10.1186/s13059-016-1010-4
- Nguyen, A., Khoo, W. H., Moran, I., Croucher, P. I., & Phan, T. G. (2018). Single cell RNA sequencing of rare immune cell populations. Frontiers in Immunology, 9. https://doi.org/10.3389/fimmu.2018.01553
- Patel, A. P., Tirosh, I., Trombetta, J. J., Shalek, A. K., Gillespie, S. M., Wakimoto, H., Cahill, D. P., Nahed, B. V., Curry, W. T., Martuza, R. L., Louis, D. N., Rozenblatt-Rosen, O., Suvà, M. L., Regev, A., & Bernstein, B. E. (2014). Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science, 344(6190), 1396-1401. https://doi.org/10.1126/science.1254257
- Baron, M., Veres, A., Wolock, S. L., Faust, A. L., Gaujoux, R., Vetere, A., Ryu, J. H., Wagner, B. K., Shen-Orr, S. S., Klein, A. M., Melton, D. A., & Yanai, I. (2016). A Single-Cell Transcriptomic Map of the Human and Mouse Pancreas Reveals Inter- and Intra-cell Population Structure. Cell Systems, 3(4), 346-360.e4. https://doi.org/10.1016/j.cels.2016.08.011
- Lähnemann, D., Köster, J., Szczurek, E., McCarthy, D. J., Hicks, S. C., Robinson, M. D., Vallejos, C. A., Campbell, K. R., Beerenwinkel, N., Mahfouz, A., Pinello, L., Skums, P., Stamatakis, A., Attolini, C. S.-O., Aparicio, S., Baaijens, J., Balvert, M., Barbanson, B. de, Cappuccio, A., … Schönhuth, A. (2020). Eleven grand challenges in single-cell data science. Genome Biology, 21(1), 31. https://doi.org/10.1186/s13059-020-1926-6
- Kharchenko, P. V., Silberstein, L., & Scadden, D. T. (2014). Bayesian approach to single-cell differential expression analysis. Nature Methods, 11(7), 740-742. https://doi.org/10.1038/nmeth.2967
- Stark, R., Grzelak, M., & Hadfield, J. (2019). RNA sequencing: the teenage years. Nature Reviews Genetics, 20(11), 631-656. https://doi.org/10.1038/s41576-019-0150-2


 Sample Submission Guidelines
Sample Submission Guidelines