We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy

Digital RNA with Perturbation of Genes (DRUG-seq) technology is a high-throughput, low-cost transcriptome sequencing method that has been extensively utilised in a variety of research scenarios, including oncology research, drug development, disease analysis and cell culture optimisation. In addition to enhancing the efficiency of drug screening, it provides more comprehensive transcriptome information for new drug development. This is expected to result in a significant enhancement in the success and efficiency of drug discovery.
DRUG-seq, otherwise referred to as high-throughput drug screening RNA sequencing, is a high-throughput platform for drug discovery. This technology was first published in the 2018 issue of Nature Communication.
The process of drug discovery is contingent upon high-throughput screening; however, contemporary platforms are limited in terms of the number of reads they can produce. RNA-seq represents a potent instrument for the investigation of drug effects through the utilisation of transcriptomic changes as proxies. Nevertheless, the standard library construction process is costly. DRUG-seq has been developed to capture the transcriptional changes detected in standard RNA-seq, at a cost that is one hundredth of the original price. In proof-of-concept experiments analysing 433 compounds at 8 doses, transcriptional profiles generated from DRUG-seq successfully grouped compounds into functional clusters based on their intended targets according to mechanism of action (MoA). Furthermore, perturbation differences reflected in transcriptome changes were detected in compounds targeting the same target, demonstrating the value of using DRUG-seq to understand target and off-target activities. Furthermore, it was demonstrated that DRUG-seq can capture both common mechanisms and differences between compound treatments and CRISPR for the same target.
DRUG-seq technology facilitates the analysis of hundreds of compounds at varying doses at the transcriptome level, enabling precise classification of diverse compounds based on their mechanism of action and intended target, as indicated by the transcriptome data they generate. Furthermore, DRUG-seq possesses extensive applications in the detection of discrepancies in transcriptome perturbations caused by different compounds targeting the same target. This capacity underscores the technology's remarkable potential to discern between target and non-target effects. Furthermore, DRUG-seq has been demonstrated to be effective in capturing differences in common mechanisms of action and transcriptome changes when comparing compound treatments targeting the same gene with CRISPR interventions.
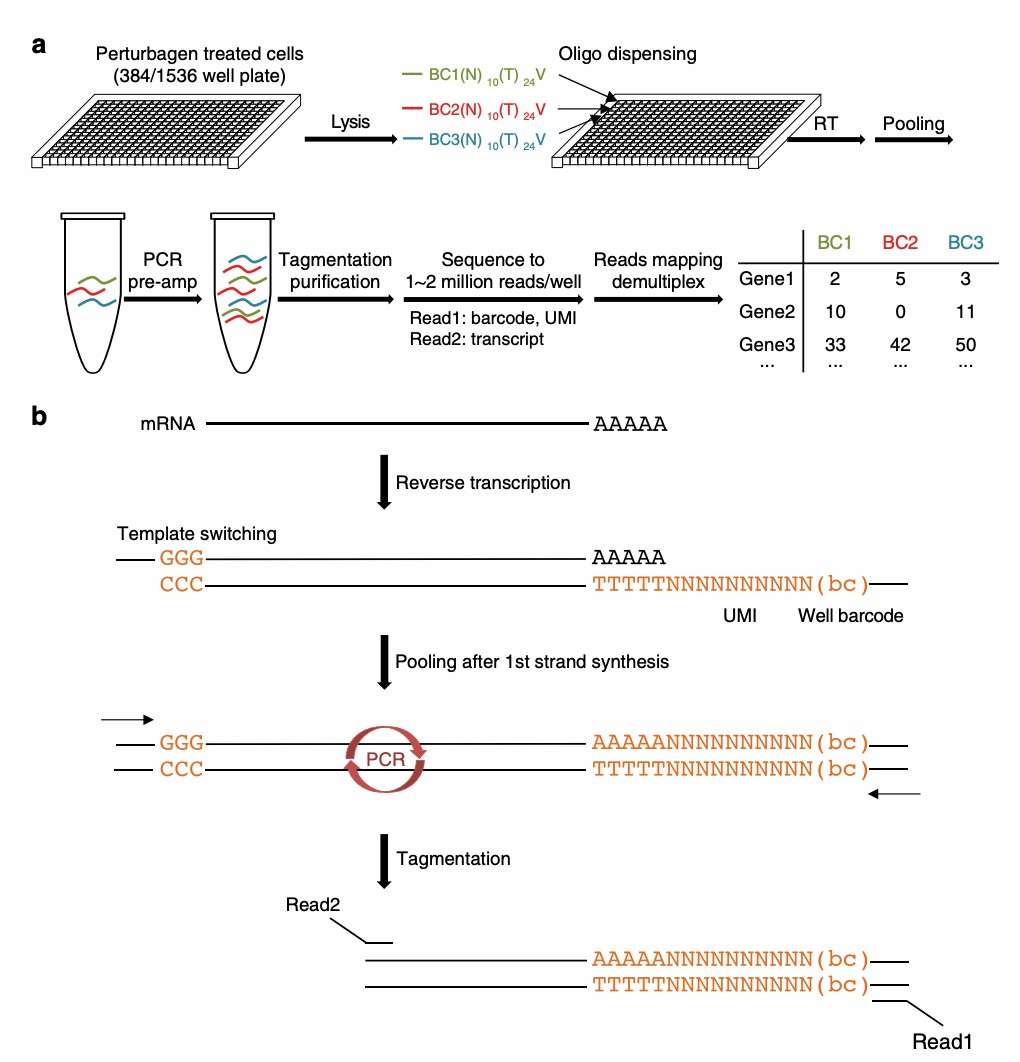 DRUG-seq overview (Ye et al., 2018)
DRUG-seq overview (Ye et al., 2018)
The optimal scenario would involve the development of a cost-effective, massively parallelised 384-well and 1536-well transcriptome analysis method that measures all genes in an unbiased manner to fully capture the transcriptional diversity induced by compounds or genetic perturbations for the purpose of drug discovery. In comparison to conventional RNA-seq, DRUG-seq necessitates only a minimal number of cells (2,000-20,000) and does not require RNA extraction. It facilitates the merging of multiple samples for library construction and sequencing, introduces molecular barcodes for precise quantification of gene expression levels, and enables concurrent large-scale sequencing. In 2022, Jingyao Li et al. published a more comprehensive pipeline for analysing DRUG-seq sequencing data in ACS Chemical Biology.
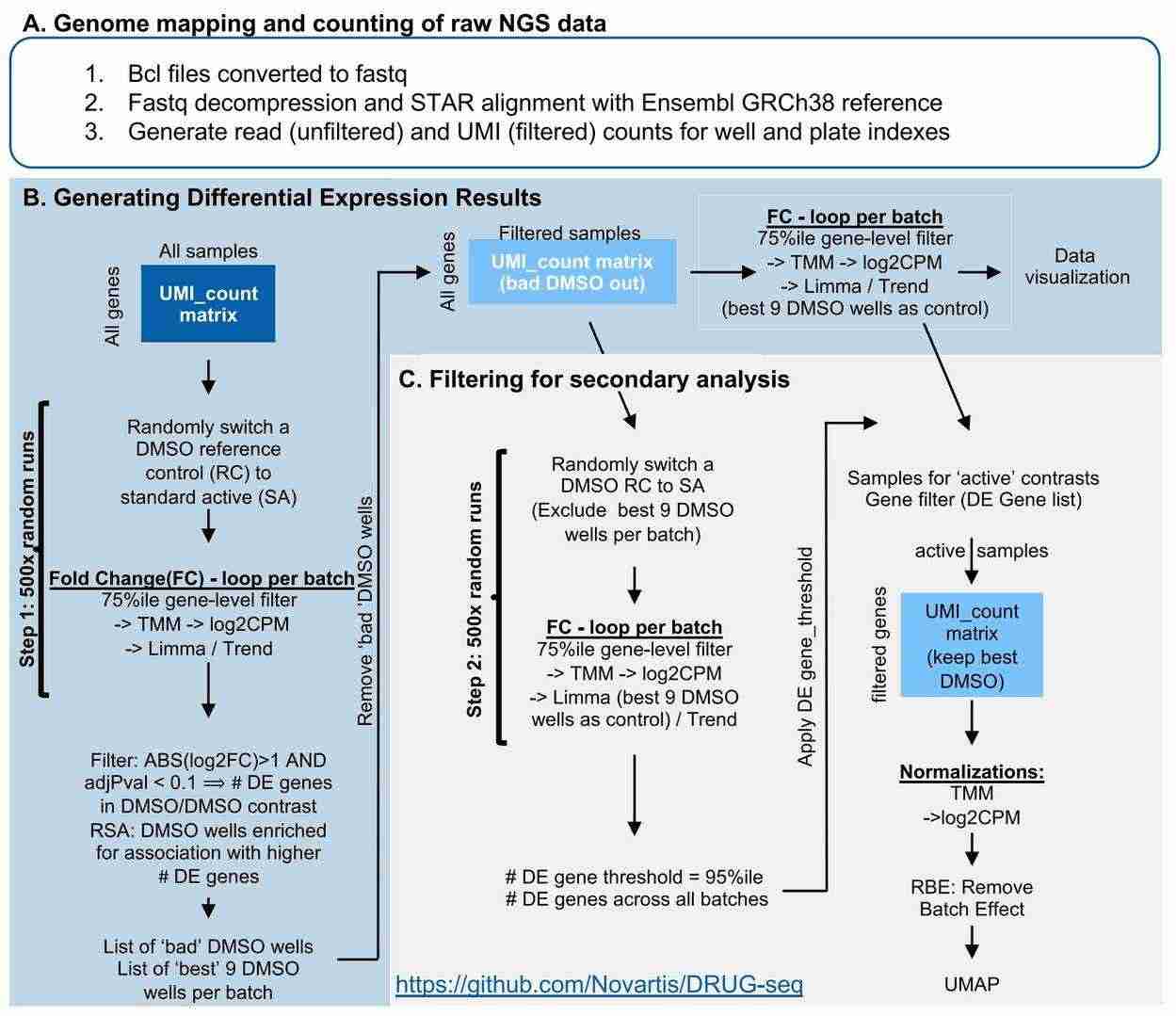 DRUG-seq analysis pipeline (Li et al., 2022)
DRUG-seq analysis pipeline (Li et al., 2022)
In summary, DRUG-seq is a powerful tool with advantages over other technologies such as RASL-seq, PLATE-seq and L1000. It is easier to perform, higher throughput, bias-free and cost-effective. DRUG-seq is a powerful tool to assist in novel compound mechanism studies, compound repurposing efforts, and the identification of genetic transcriptional networks using CRISPR/CAS9-based gene knockouts.
Take the Next Step: Explore Related Services
Venous malformations (VM) represent the most prevalent form of vascular malformation, arising from somatic mutations within venous endothelial cells (VECs). The prevalence of VM ranges from 1 in 10,000 to 2 in 10,000 individuals. The clinical manifestations associated with VM include structural abnormalities, discomfort, swelling, and bleeding at the affected site, potentially leading to organ dysfunction. Current treatment options include sclerotherapy and surgical resection, which are effective in controlling the disease, but are not curative, are expensive, and carry high surgical risks.
A significant lacuna persists in pharmacological treatment options for VM, with a paucity of FDA-approved drugs currently available, primarily due to the absence of cost-effective disease models capable of reproducing symptoms and facilitating large-scale replication. The causative somatic mutations of VM are typically non-heritable and localised in their onset, rendering conventional transgenic animal breeding methods inapplicable. Furthermore, the utilisation of AAV viruses for the delivery of mutants in mouse models, while exhibiting the potential to induce vascular malformations, is encumbered by substantial expenses, suboptimal stability, and the presence of significant human-mouse discrepancies. Consequently, the establishment of a novel vascular malformation model is imperative for the advancement of our understanding of the pathological mechanisms underpinning these malformations and for facilitating the development of effective pharmaceutical interventions.
Recently, Kai Wang's team at Peking University recently published a research paper in Cell Stem Cell entitled 'Generation of iPSC-derived human venous endothelial cells for the modelling of vascular malformations and drug discovery'. The team developed an efficient differentiation protocol for venous endothelial cells (iVEC), and further reproduced the pathological features of venous malformations using iVEC derived from induced pluripotent stem cells (iPSCs) carrying the TIE2-L914F heterozygous mutation. The combination of deep learning and DRUG-seq technology led to the discovery that Bosutinib, an FDA-approved leukaemia drug, significantly improved the pathological phenotype of venous malformations. The utilisation of iPSC-derived multiple disease models, in conjunction with the pioneering application of DRUG-seq technology in drug screening, has the potential to overcome the challenges posed by vascular malformations and open new therapeutic pathways.
Main Research Content
The present study focuses on the most common pathogenic mutation in VMs, TIE2 p.L914F (c.2740 C>T).TIE2 (TEK) is a member of the receptor tyrosine kinase subfamily that is expressed predominantly in the endothelium, and plays an important role in angiogenesis, remodelling, maturation and integrity. The most widely used cellular model for simulating vascular malformations is the overexpression of mutants of HUVECs. Nevertheless, a number of significant distortions have been identified in this class of models. Firstly, HUVECs expanded in vitro lose some of their venous properties. Secondly, overexpression invariably produces a higher dose of mutants than heterozygous mutations in the patient.
In order to more accurately replicate the characteristics of veins, the research team developed a robust differentiation scheme for iVECs. This scheme involves the modulation of the retinoic acid signalling pathway to regulate the cell cycle, thereby promoting specific differentiation towards the venous endothelium. In order to address the mutation copy number issue, the team introduced the L914F heterozygous point mutation into the TIE2 gene of iPSCs using gene editing techniques.
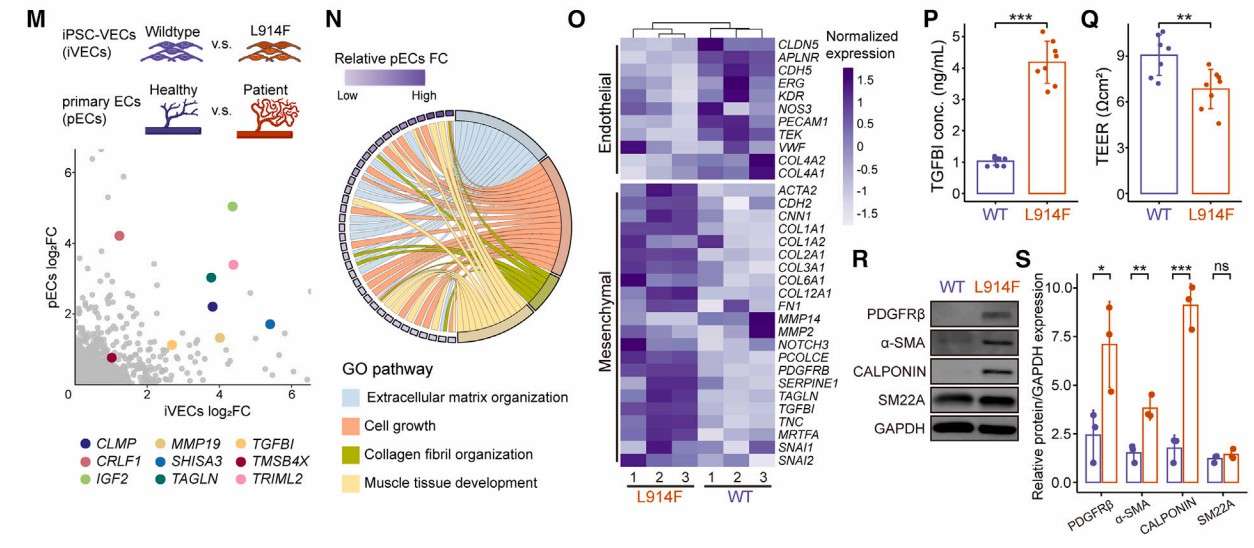 TIE2-L914F-mutated iVECs phenocopied VMs in vitro (Pan et al., 2025)
TIE2-L914F-mutated iVECs phenocopied VMs in vitro (Pan et al., 2025)
Following a comprehensive review of the extant literature, the investigators established a reliable and effective venous endothelial differentiation protocol. This protocol incorporated vascular endothelial growth factor (VEGFA), fibroblast growth factor (FGF2), NOTCH inhibitor (DAPT), and retinoic acid (RA) as its principal components, with continuous adjustments being made to the differentiation conditions. Subsequent studies have elucidated that RA, among other factors, can induce an early blockade of G1, thereby promoting differentiation into the venous spectrum.
A comparison of single-cell sequencing data from iVECs and iAECs suggests that MEF2C may promote venous endothelial differentiation, a hypothesis that has not previously been put forward. The researchers overexpressed the MEF2C gene and found that the proportion of venous endothelial cells was increased. Furthermore, the functional validation of the iVECs demonstrated that they exhibit venous characteristics, including low levels of nitric oxide production, a weak response to shear stress, and an increased ability to recruit immune cells.In vivo transplantation of iVECs also consistently expressed venous markers and formed a perfusable lumen.
Subsequently, the investigators introduced the TIE2-L914F heterozygous mutation into iPSCs using CRISPR-Cas9. The mutant iVECs recapitulated the pathological phenotype of VM, including disturbed F-actin, enhanced proliferative and anti-apoptotic capacity, and diminished lumen-forming capacity. Bulk RNA-seq supported the abnormalities in the corresponding pathways and gene expression levels in the mutant VM. Further transplantation of mutant iVECs under the renal capsule in mice resulted in the formation of aberrant vessels and produced significant functional abnormalities.
In order to identify a suitable pharmaceutical agent for the treatment of VM, the research team employed a deep learning-based DLEPS method for drug prediction.In conjunction with DRUG-Seq, the team hypothesised that Bosutinib is a potentially available pharmaceutical agent.Administration of the drug to the mutant group resulted in a significant improvement in their abnormal VM phenotype, thereby demonstrating the therapeutic effect of Bosutinib. Further exploration of the genomic and proteomic expression of the administered group revealed that Bosutinib inhibited cell proliferation by attenuating the level of endothelial mesenchymal transition in the mutant group, which in turn alleviated the aberrant phenotype caused by the TIE2-L914F mutation.
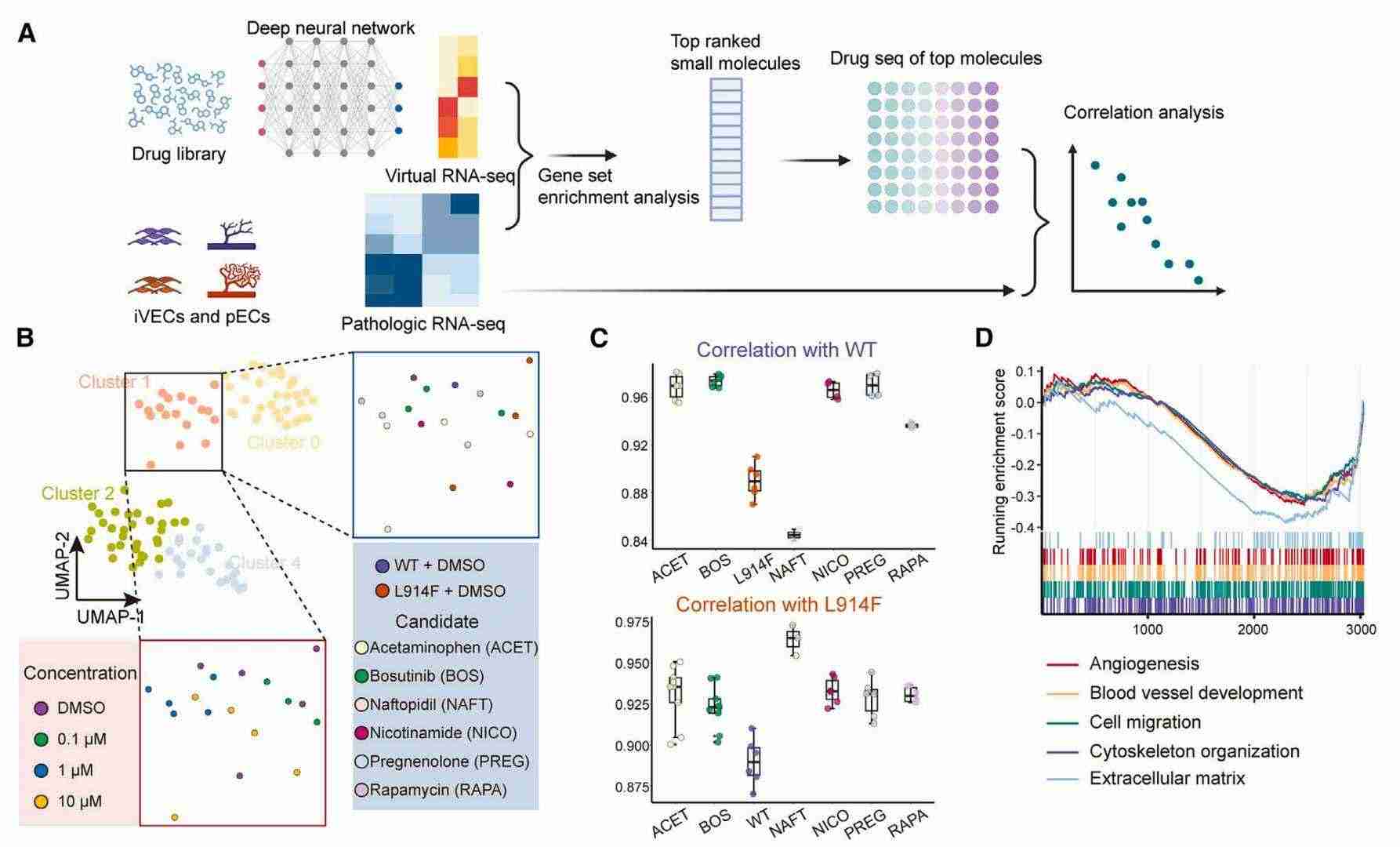 AI-supported discovery of bosutinib to alleviate VMs (Pan et al., 2025)
AI-supported discovery of bosutinib to alleviate VMs (Pan et al., 2025)
The present study identified the genes required for retinoic acid RA-mediated cellular G1 phase block and the subsequent activation of progenitor cells for transformation to venous endothelial cells. The authors introduced the L914F heterozygous mutation in iPSCs and differentiated them into venous endothelium, which serves as a model for venous malformations. They identified Bosutinib as effective in rescuing the disease phenotype in vitro and in vivo using deep learning and high-throughput DRUG-Seq methodology. This opens up more possibilities for potential drug discovery and clinical therapeutic targets in the field of vascular malformations.
The utilisation of iPSC-derived vascular malformation models in conjunction with DRUG-seq has resulted in a substantial acceleration in the development process of novel therapeutic drugs. Subsequent studies could encompass the exploration of DRUG-seq technology's application in alternative disease models, along with the incorporation of enhanced bioinformatics analyses. This would facilitate a more profound comprehension of drug mechanisms of action and potential off-target effects.
References

CD Genomics is transforming biomedical potential into precision insights through seamless sequencing and advanced bioinformatics.
We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy