
- Home
- Solutions
- Epigenome Sequencing
- Metagenomics Sequencing
- DRUG-seq Service


DRUG-se represents a cutting-edge high-throughput drug screening platform based on RNA sequencing. This technology is designed for transcriptomic profiling from minimal cell quantities in 96- or 384-well plates. It utilizes well barcoding and molecular barcodes quantification to enable full-length transcript amplification. DRUG-seq constitutes a crucial step in the early stages of drug candidate selection during Chemistry, Manufacturing, and Controls (CMC) submission. This methodology allows for the simultaneous detection of transcriptomes across all samples in large-scale high-throughput screening, involving hundreds of candidate compounds and experimental conditions. The resulting data provides comprehensive insights into drug mechanisms of action.
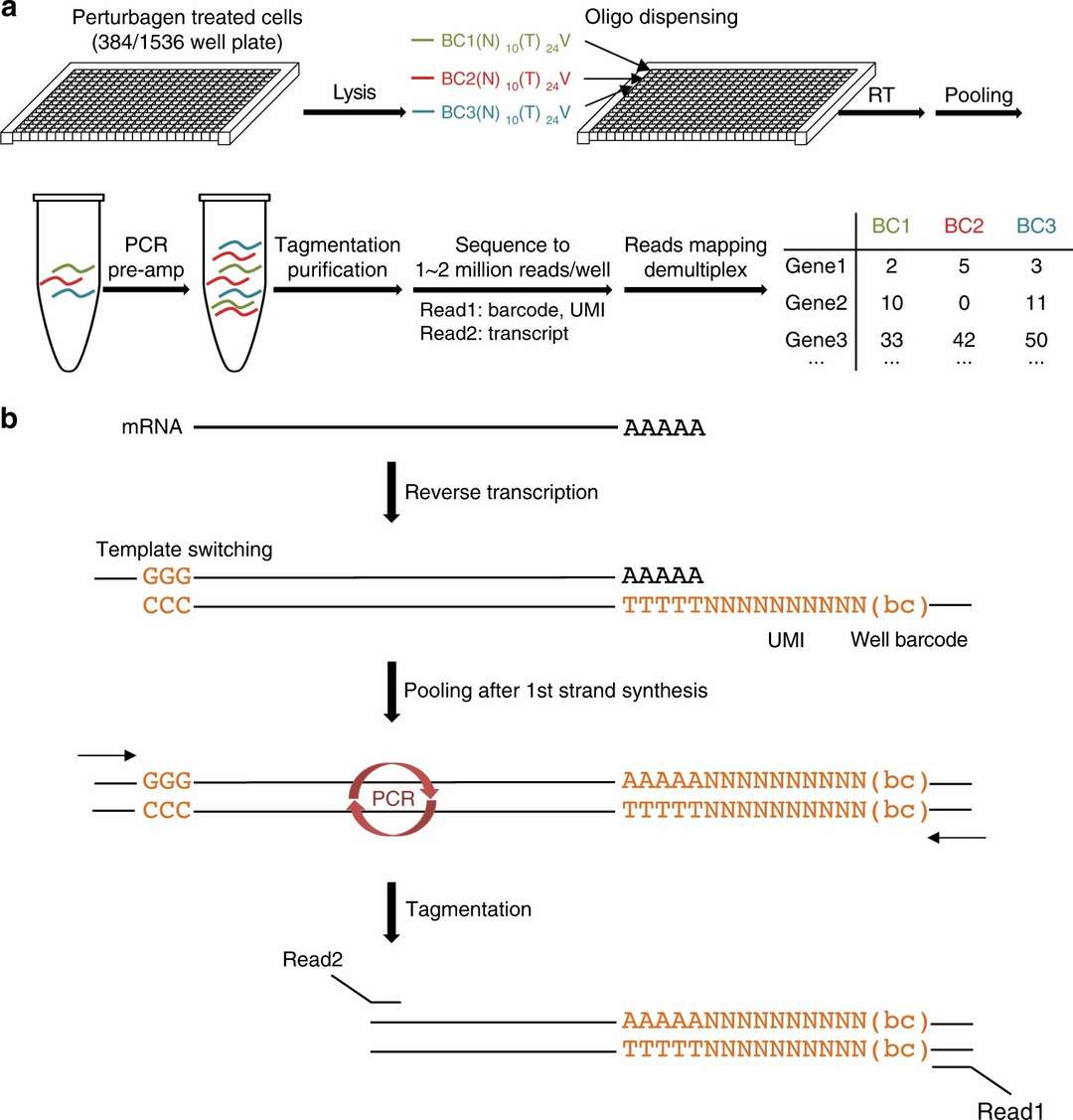 Figure 1. DRUG-seq overview.
Figure 1. DRUG-seq overview.
The advent of high-throughput sequencing technologies has markedly transformed the field of pharmacological research, especially in the identification and validation of novel therapeutic compounds. Within this spectrum, DRUG-seq has emerged as a cornerstone technique, effectively merging the comprehensive capabilities of full-length RNA sequencing with the scalability essential for industrial-level drug screening. This methodology facilitates the precise quantification of gene expression, thereby allowing researchers to identify specific metabolic pathways influenced by drug treatments.
In the realm of drug discovery, it is crucial to evaluate the transcriptional changes induced by compounds at the cellular level. DRUG-seq fulfills this requirement by enabling the simultaneous transcriptomic profiling of numerous candidate compounds within a single experiment. Furthermore, the technology is adaptable to a variety of experimental conditions, encompassing different cell types, drug dosages, and time points. By offering a detailed perspective on how compounds modulate gene expression, DRUG-seq significantly contributes to the identification of promising drug candidates and enhances the probability of success in subsequent drug development stages.
Sequencing Platform: Illumina NovaSeq
Data Output: 1–2 GB per well
Additional tailored bioinformatic analyses are available upon request, providing a more specific investigation into particular drug effects, pathways, or gene interactions. This can include advanced statistical modeling, in-depth pathway interrogation, or integration with other omics data types for a multi-layered approach to understanding drug efficacy and mechanism.
| Sample Type | Sample Quantity | Handling Instructions | Shipping Method |
|---|---|---|---|
| Adherent Cells | 2,000–10,000 cells | 1. Wash cells thoroughly twice with 50–100 μL of phosphate-buffered saline (PBS). 2. After complete removal of PBS, add 30 μL of Lysis Mix. 3. Gently pipette to resuspend and lyse cells at room temperature for 6 minutes. |
1. Freeze in liquid nitrogen, followed by storage at -80°C. 2. Ship on dry ice. 3. Avoid freeze-thaw cycles; ship samples on the same day as processing. |
| Suspended Cells | 2,000–10,000 cells | 1. Mix the cell culture suspension thoroughly by pipetting. 2. Transfer 10 μL from each well, mixing with an equal volume of Lysis Mix (10 μL). 3. Allow to lyse at room temperature for 6 minutes. |
1. Freeze in liquid nitrogen, followed by storage at -80°C. 2. Ship on dry ice. 3. Avoid freeze-thaw cycles; ship samples on the same day as processing. |
Notes
DRUG-seq and RNA-seq are both powerful techniques used to study gene expression at the transcriptome level, but they serve distinct purposes and offer different advantages in specific contexts. Below is a detailed comparison of the two methods:
| Aspect | DRUG-seq | RNA-seq |
|---|---|---|
| Purpose | High-throughput drug screening via transcriptome profiling | Comprehensive transcriptome analysis for various biological applications |
| Throughput | Allows screening of 96- or 384-well plates simultaneously | Typically lower throughput, focusing on individual samples |
| Cost | More cost-efficient due to pooled sample sequencing | Higher cost per sample, especially in high-throughput settings |
| Starting Material | Requires a small number of cells (2,000–20,000 cells per sample) | Variable, but typically requires a higher starting material depending on protocol |
| Time Efficiency | Short experimental cycle with simultaneous multi-sample processing | Longer experimental cycles, with samples processed individually |
| Resolution | Focuses on gene-level resolution, particularly for drug effects | Provides full transcriptome coverage but less focused on drug-specific effects |
| Application Focus | Drug discovery, mechanism-of-action studies, off-target effect evaluation | Broad applications: disease research, gene expression analysis, differential expression studies |
| Data Analysis | Supports both standardized and customized analyses for specific drug effects | Standard RNA-seq analysis pipelines for general transcriptomic data |
| Sensitivity | High sensitivity to changes in gene expression related to drug action | High sensitivity to detect changes across the entire transcriptome |
| Functional Insight | Specifically designed to reveal drug-induced transcriptional changes and metabolic pathway alterations | Provides a holistic view of the transcriptome, not specific to drug-related changes |
| Bioinformatics Requirements | Integrates with bioinformatic pipelines that emphasize drug-induced expression changes | Requires more generalized bioinformatic tools, adaptable to various research questions |
Validation of Compound Mechanisms of Action Using DRUG-seq Technology
In a study involving the validation of 433 compounds across eight different dosages, transcriptomic profiles obtained through the DRUG-seq technology successfully facilitated the functional classification of these compounds based on their anticipated mechanisms of action (MoAs). Significant variations in transcriptional alterations were detected among compounds targeting the same biological entities, providing evidence for the utility of DRUG-seq in elucidating both on-target and off-target activities of compounds.
The findings underscore the capacity of DRUG-seq to differentiate between the perturbations in transcriptomic responses associated with distinct compounds, thereby contributing to a deeper understanding of the complex biological interactions inherent to pharmaceutical agents. This capability enhances the assessment of therapeutic potential and supports the optimization of drug discovery processes.
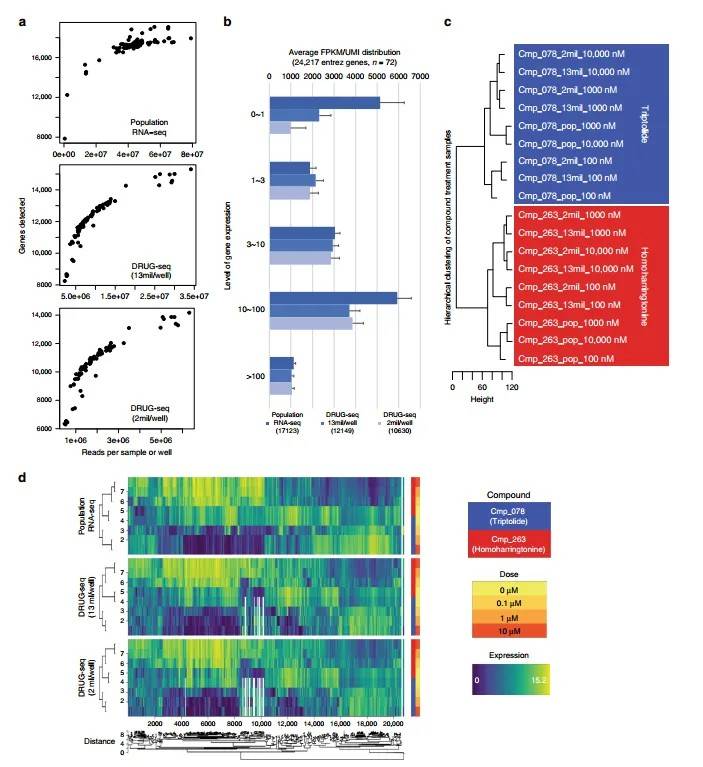 Figure 2 illustrates the comparable performance of DRUG-seq and standard RNA-seq.
Figure 2 illustrates the comparable performance of DRUG-seq and standard RNA-seq.
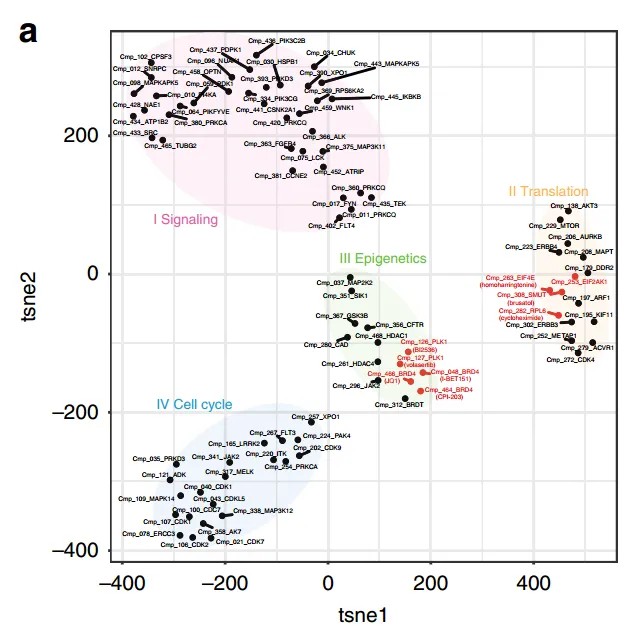 Figure 3 presents the t-SNE clustering analysis of compounds and their regulatory mechanisms.
Figure 3 presents the t-SNE clustering analysis of compounds and their regulatory mechanisms.
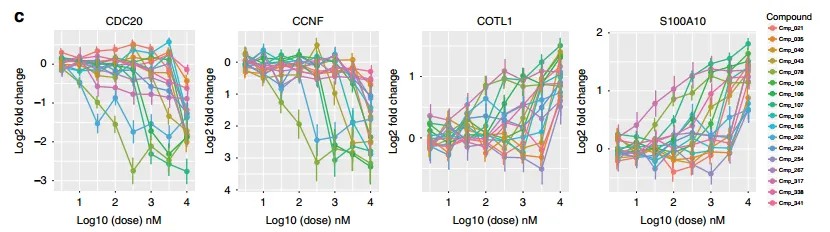
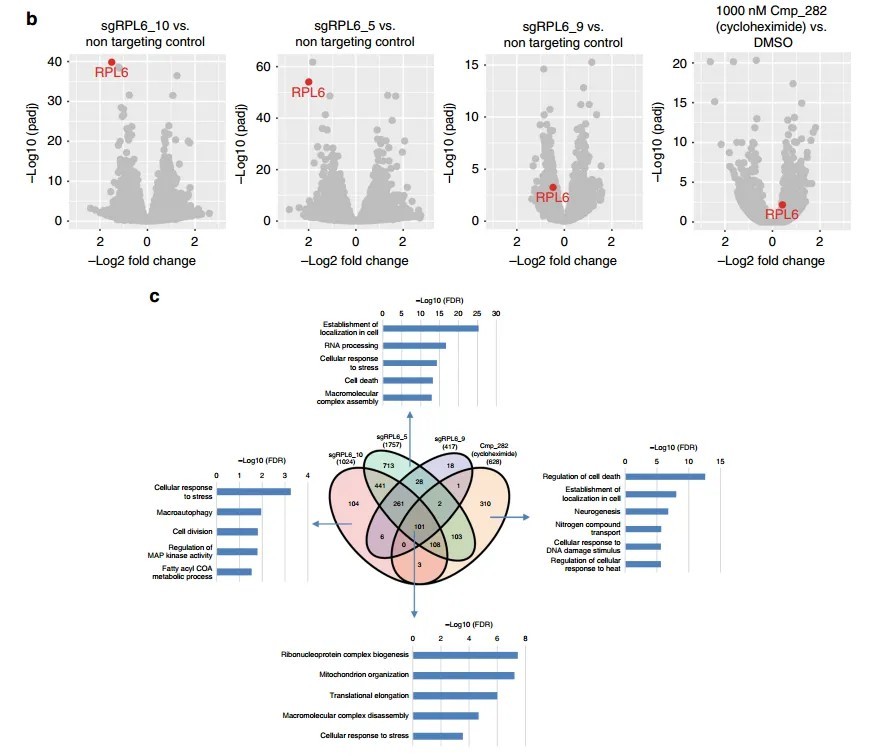
Figure 4 highlights the expression changes of specific genes following treatment with varying compound concentrations.
Figure 5 depicts the differential gene expression analysis post-compound treatment.
References

CD Genomics is transforming biomedical potential into precision insights through seamless sequencing and advanced bioinformatics.