What is Animal/Plant Exome Sequencing
Exon capture sequencing is a method used to extract and sequence exons (collections of all exons) in the genome and obtain exon variation in a single organism sample. This approach allows research to quickly focus on the parts of the genome that are most likely to influence phenotypic variation. Compared with whole genome sequencing, the method of exon capture sequencing simplifies the genome of target species, greatly reduces redundant sequences, can more accurately and quickly locate candidate genes related to specific traits, effectively reduces sequencing costs, and makes it widely used in animal and plant research.
Animal and plant exome sequencing plays a crucial role in various fields, including genetics, disease diagnostics, evolutionary biology, and agricultural breeding. By precisely targeting protein-coding regions, this approach enables researchers to identify genetic variations associated with key traits, providing insights into hereditary diseases, adaptive evolution, and species diversity. In agriculture, exome sequencing accelerates genomic selection, aiding in the breeding of crops and livestock with desirable characteristics such as disease resistance and high yield. Furthermore, it supports conservation efforts by analyzing genetic diversity and resilience in endangered species.
Animal and Plant Whole Exome Series Products
Based on hybridization capture sequencing technology and nucleotide synthesis platform, targeted sequencing of exons of animals and plants can be widely used in molecular breeding, population genetics analysis, BSA fine mapping, mutant library sequencing, etc.
| Product Name | Reference Genome | Target Region Size |
|---|---|---|
| Wheat Whole-Exome Capture Product | Triticum aestivum (IWGSC V2.1) | 132.6 Mb |
| Barley Whole-Exome Capture Product | Hordeum vulgare (MorexV3) | 42.0 Mb |
| Masson Pine Whole-Exome Capture Product | Pinus tabuliformis | 111.1 Mb |
| Maize Whole-Exome Capture Product | Zea mays (B73 V5) | 45.5 Mb |
| Mouse Whole-Exome Capture Product | Mus musculus (mm39) | 38.4 Mb |
| Rat Whole-Exome Capture Product | Rattus norvegicus (GRCr8) | 38.3 Mb |
| Dog Whole-Exome Capture Product | Canis lupus familiaris (canFam4) | 36.0 Mb |
| Pig Whole-Exome Capture Product | Sus scrofa (Sscrofa11.1) | 35.8 Mb |
| Cattle Whole-Exome Capture Product | Bos taurus (ARS-UCD2.0) | 36.9 Mb |
| Chicken Whole-Exome Capture Product | Gallus gallus (GRCg7b) | 32.1 Mb |
CD Genomics provides comprehensive exome sequencing services for a wide range of animal and plant species. By utilizing advanced exon capture technology and next-generation sequencing (NGS) platforms, we ensure high-quality data for identifying key genetic variations.
Advantages of Our Animal/Plant Exome Sequencing Service
High Specificity: Targeted sequencing of exonic regions or specific SNP sites of interest.
Enhanced Efficiency: Utilizes next-generation sequencing (NGS) platforms for faster and more efficient sequencing.
High Accuracy: Deep sequencing coverage ensures precise and reliable results.
Cost-Effective: Focuses only on target regions, significantly reducing research costs.
Applications of Animal/Plant Exome Sequencing
Applications of Exome Sequencing in Plants:
Crop Improvement: Exome capture technology has been used to identify gene variations related to yield, disease resistance, and adaptability in crops like soybean, rice, and wheat. It also aids in constructing genetic maps and QTL mapping.
Microexon Research: Microexons in plants play significant roles in gene expression regulation, development, and disease processes.
Complex Genome Analysis: In complex genome plants, exome capture significantly reduces sequencing costs and enhances data quality, especially in the absence of a reference genome.
Applications of Exome Sequencing in Animals:
Disease Research and Genetic Research: Exome sequencing is widely used in animal disease research and genetic studies, such as identifying gene variations associated with pheochromocytoma in canines and pathogenic mutations in rare human diseases.
Breeding and Evolution Studies: Exome sequencing helps identify gene variations related to productive traits, accelerating genetic improvement in animal breeding.
Microexon Research: Microexons in animals are involved in gene expression regulation and various biological processes.
Animal/Plant Exome Sequencing Workflow
Exome sequencing involves extracting and preprocessing DNA, capturing target exonic regions using specific probes, enriching and amplifying the captured fragments, sequencing them with platforms like Illumina, and analyzing the data by aligning it to a reference genome to identify variants that could impact protein function.

Service Specifications
| Sample Requirements
Sample Requirements:
|
|
Sequencing
|
|
| Bioinformatics Analysis
We provide multiple customized bioinformatics analyses:
|
Analysis Pipeline
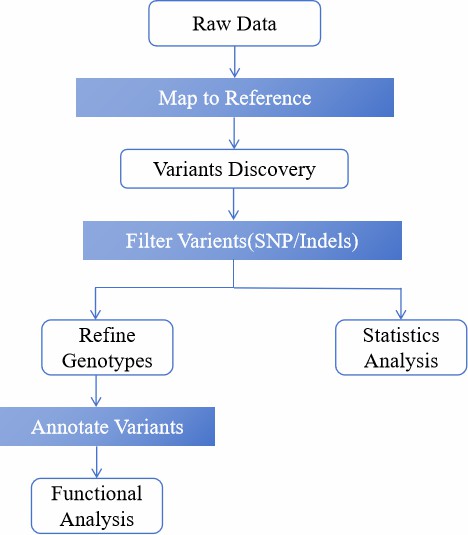
Deliverables
- The original sequencing data
- Experimental results
- Data analysis report
- Details in Whole Exome Sequencing for your writing (customization)
CD Genomics offers comprehensive exome sequencing services for both animal and plant species, providing a complete solution from sample preparation to advanced bioinformatics analysis. Our service includes exonic region capture, high-coverage sequencing, and accurate variant identification, tailored to meet your specific research needs. We utilize state-of-the-art exon capture technology and next-generation sequencing platforms to ensure high-quality results. Whether you are studying crop improvement, disease diagnostics, or genetic diversity, our animal and plant exome sequencing services deliver precise, cost-effective insights. For further information or to discuss custom requirements, feel free to reach out to us.
Partial results are shown below:

Raw data filtering results.

Sequencing error rate distribution.

GC content distribution of samples.

Sequencing depth.

SNV identification result.
1.How to choose the appropriate plant and animal exome sequencing platform?
When selecting a sequencing platform, the following factors should be considered:
Target region size: For example, the exome of wheat contains approximately 170-340 Mb, whereas the human exome is about 30 Mb.
Probe design: Array-based capture is suitable for large-scale studies, while liquid-phase capture is more appropriate for small sample sizes or complex genomes.
Sequencing depth: The required coverage depth should be determined based on the research objectives. For instance, higher coverage depth is typically needed for disease diagnosis.
2.What are the future development trends of plant and animal exome sequencing?
With advancements in sequencing technology, plant and animal exome sequencing will become more efficient and cost-effective in the future. For example:
Multispecies Comparative Analysis: By integrating exome data from different species, broader evolutionary and functional relationships can be revealed.
Single-Cell Sequencing: Combining single-cell technologies to study genetic heterogeneity between individual cells.
AI-Assisted Analysis: Utilizing machine learning algorithms to enhance the efficiency and accuracy of data analysis.
3.What is the advantage of exome sequencing over other genomic methods?
The primary advantages of exome sequencing are its lower cost compared to whole genome sequencing and its focus on exons, which are the most functionally significant regions of the genome. This makes it particularly useful for identifying mutations that affect gene function.
4.How do you analyze the data from exome sequencing?
Exome sequencing data is analyzed using bioinformatics tools that align the sequence reads to a reference genome, identify variants (such as single nucleotide polymorphisms or indels), and perform functional annotation to predict the potential impact of these variants.
Efficient Genome-Wide Detection and Cataloging of EMS-Induced Mutations Using Exome Capture and Next-Generation Sequencing
Journal: Nature Biotechnology
Impact factor: 33.1
Published: Feb 2019
Background
With the domestication and intensive breeding of crops, the diversity of disease resistance genes has gradually decreased, making crops more susceptible to disease threats. Although wild relatives often harbor multiple disease resistance genes, traditional R-gene cloning methods, such as positional cloning and mutagenesis genomics, are typically difficult to apply to these genes due to the presence of undesirable agronomic traits. Especially in wild species, identifying a single disease resistance gene and incorporating it into a susceptible background often requires several generations of selection, making the gene discovery process highly cumbersome.
Resluts
In this study, the authors selected the diploid ancestral species of the wheat D genome, Aegilops tauschii, as the research subject. This species exhibits strong resistance to wheat stem rust pathogens and is a valuable source of wheat stem rust resistance genes. The authors collected 174 samples of Ae. tauschii ssp. Strangulata and selected 21 samples of Ae. tauschii ssp. Tauschii as outgroup materials, with the already cloned R genes Sr33 and Sr45 serving as positive controls. The authors designed capture probes targeting the genomic regions of the NLRs in Ae. tauschii and 317 SNP loci that are evenly distributed across the genome. Using deep sequencing, they obtained 249–336 full-length NLR genes and 1,312–2,170 non-full-length NLR genes in each sample, and identified variations within the target regions. Combining the disease resistance data from 151 samples, the authors conducted a K-mer-based candidate gene association analysis and identified multiple Sr genes within Ae. tauschii ssp. Strangulata, including the already cloned Sr33, Sr45, Sr46, and SrTA1662 genes. Previous wheat studies have revealed that these four genes have introgressed from the Ae. tauschii subgenus into common wheat.
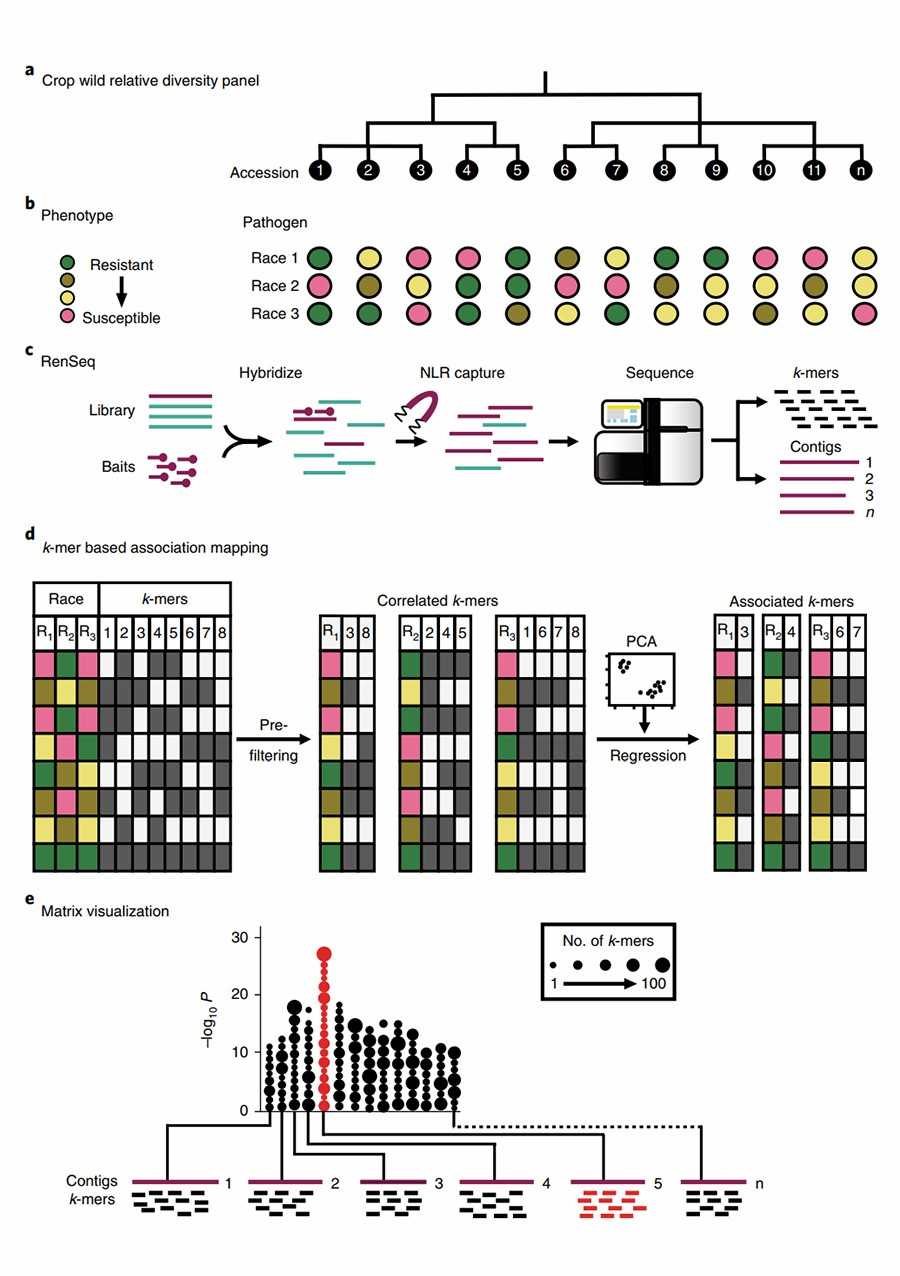 Figure1.Combining association genetics and R gene enrichment sequencing (AgRenSeq) for R gene cloning.(Arora, S.et.al,2019)
Figure1.Combining association genetics and R gene enrichment sequencing (AgRenSeq) for R gene cloning.(Arora, S.et.al,2019)
In subsequent breeding processes, these resistance genes can be introgressed into the wheat genome through tracking of target genes, thereby enhancing wheat resistance.
Reference:
- Arora, S., Steuernagel, B., Gaurav, K. et al. Resistance gene cloning from a wild crop relative by sequence capture and association genetics. Nat Biotechnol 37, 139–143 (2019). https://doi.org/10.1038/s41587-018-0007-9
Here are some publications that have been successfully published using our services or other related services:
Combinations of Bacteriophage Are Efficacious against Multidrug-Resistant Pseudomonas aeruginosa and Enhance Sensitivity to Carbapenem Antibiotics
Journal: Viruses
Year: 2024
Distinct functions of wild-type and R273H mutant Δ133p53α differentially regulate glioblastoma aggressiveness and therapy-induced senescence
Journal: Cell Death & Disease
Year: 2024
Genomic transmission clusters and circulating lineages of Mycobacterium tuberculosis among refugees residing in refugee camps in Ethiopia
Journal: Infection, Genetics and Evolution
Year: 2023
A de novo assembly of genomic dataset sequences of the sugar beet root maggot Tetanops myopaeformis, TmSBRM_v1.0
Journal: Data in Brief
Year: 2024
Sex hormones, sex chromosomes, and microbiota: Identification of Akkermansia muciniphila as an estrogen-responsive microbiota
Journal: Microbiota Host
Year: 2024
See more articles published by our clients.


 Sample Submission Guidelines
Sample Submission Guidelines
