Since Nature published the complete map of the human X chromosome, the high-quality telomere-to-telomere (T2T) genome, obtained by combining various sequencing technologies such as PacBio HiFi, ONT Ultra-long, and Hi-C, has brought new opportunities to genome assembly research. To facilitate targeted learning, a summary of the recently published T2T genome articles has been made, outlining the overall analysis approaches of different species.
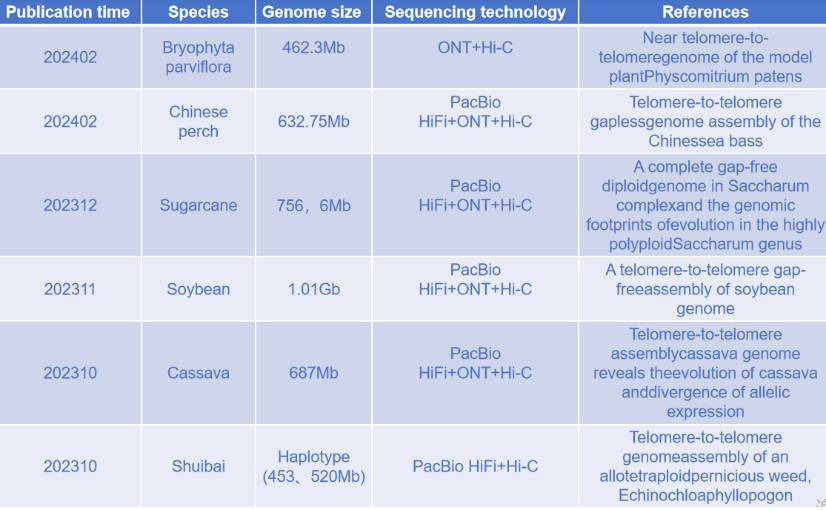 Summary of T2T genome in recent research
Summary of T2T genome in recent research
T2T Genome of Physcomitrium Patens

- Date of publication: February 2024
- Journal: Nature Plants (IF=17.352)
- Sequencing strategy: ONT+Hi-C
- Main research ideas:
In this paper, ONT reads was used as the initial skeleton to assemble NextDenovo, and it was mounted on the chromosome with Hi-C technology. The final genome was 473Mb, including 26 chromosomes with 23 gaps and two telomere sequences (P. patens V6 genome).
By using the RNA-seq data set in the database, two annotated versions were integrated to assist the annotation of the new genome structure of P. patens V6. Hi-C sequencing identified topological correlation domains (TADs) across chromosomes. The distribution of A/B interval confirmed that the chromosomes of P. patens showed more uniform distribution of eukaryotic chromatin and heterochromatin than that of many flowering plants.
The distribution of LTR family in the genome was determined by principal component analysis. Combined with the results of ChIP-seq, it was proved that there were only 26 chromosomes in P. patens, which further indicated that chromosomes could be divided into three types: centromere type, subcentromere type and secondary centromere type.
A remarkable structure (GGCCCA)n was observed in the new telomere structure, which is different from the traditional (TTTAGGG)n repetition and may be the result of the secondary telomere structure in the process of chromosome breakage and healing. Combined with genome-wide repeat analysis, the effectiveness of p.paterens in repairing DNA double-stranded breaks was revealed. When the DNA double-strand break repair mechanism can't repair the broken chromosome, it will start the chromosome healing process, eventually form new telomeres and lead to the emergence of new chromosomes.
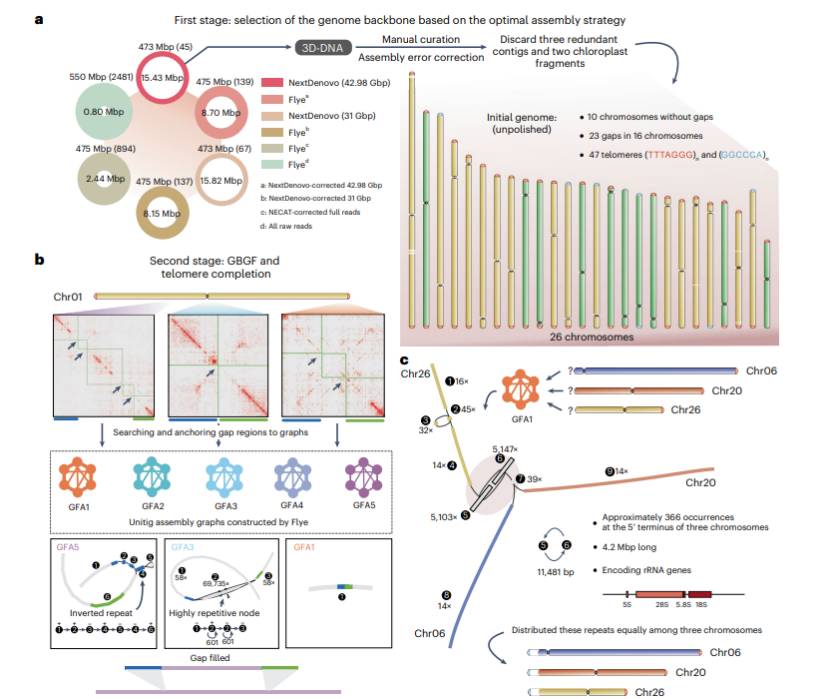 T2T Assembly of Physcomitrium patens V6 Genome (Bi et al., 2023)
T2T Assembly of Physcomitrium patens V6 Genome (Bi et al., 2023)
Service you may interested in
T2T Genome of Maize

- Date of publication: June 2023
- Journal: Nature Genetics(IF=30.8)
- Sequencing strategy: PAC Biohifi+ONT+Hi-C.
- Main research ideas:
In this paper, Mo17 inbred lines were selected as materials, using PacBio HiFi sequencing data as the skeleton, ONT Ultralong data as a supplement to Gap, and combining with Hi-C sequencing technology, the T2T genome of maize with 10 chromosomes and a genome size of 2,178.6 Mb was obtained. 1,029 new genes were discovered and corresponding annotation information was obtained.
ChIP-seq technology verified the genome structure of 10 centromeres, and 82 genes were identified in Mo17 centromeres, of which 72 had homologous genes in maize NAM inbred lines and 52 had homologous genes in sorghum. Centromere gene pairs tended to be tissue-specific.
The comparative analysis of telomere and sub-telomere lengths of 10 chromosomes shows that the average length of 20 telomeres in the complete T2T Mo17 genome is 26.1 kb, and each telomere has about 3,700 telomere duplicate copies. The G-rich telomere repeat chain (5'-TTTAGGG-3') always faces the ends of all ten chromosomes. The analysis of 20 fully assembled chromosome ends revealed the extensive length and sequence composition changes of sub-telomere regions.
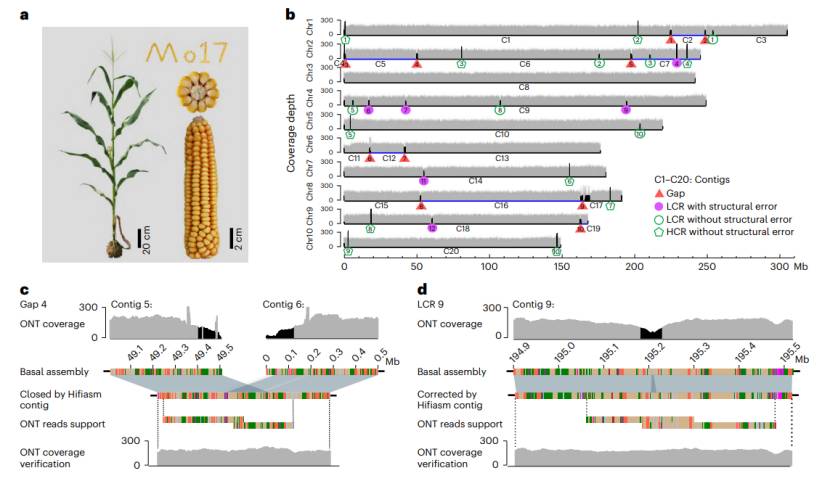 T2T assembly of Mo17 maize genome (Chen et al., 2023)
T2T assembly of Mo17 maize genome (Chen et al., 2023)
Combining the above two cases, we can summarize the research ideas of plant T2T genome:
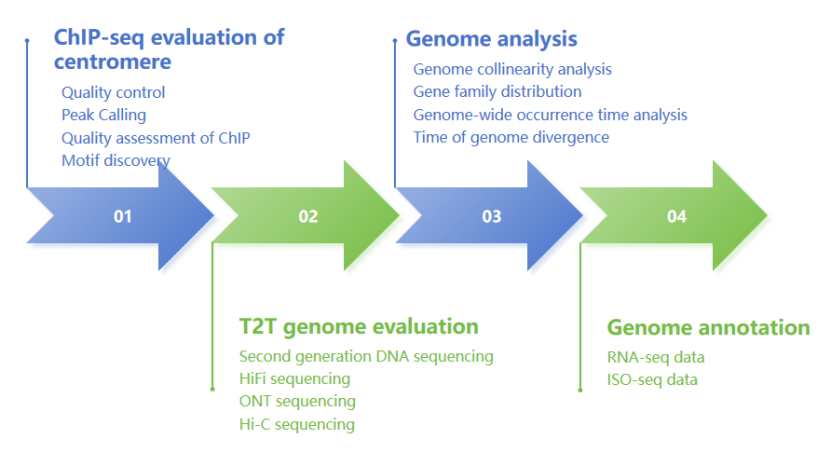 Workflow of T2T genome analysis
Workflow of T2T genome analysis
Through the above research ideas, we can find that the high-quality genome skeleton obtained based on PacBio HiFi, combined with ONT. Ultra-long sequencing supplemented gap and Hi-C technology further improved the integrity of genome assembly, and finally reached T2T level. T2T overcomes the difficulty in assembling centromeres or high-repetitive regions, greatly improves the continuity and integrity of chromosomes, and is helpful to further study the sequence or structure of high-repetitive regions in the genome. T2T genome assembly is helpful to further explore new genes and understand the structure of centromere region, and at the same time, it is also helpful to deeply study the methylation level, repetitive sequence variation, transposon movement and centromere evolution of the whole genome.
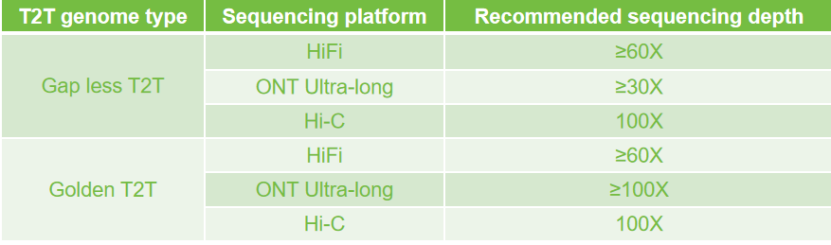 Recommended different T2T sequencing strategies
Recommended different T2T sequencing strategies
Tips for T2T Genome Detection
- The second and third generation DNA sequencing nucleic acids are preferably the same tissue of the same individual.
- It is best to set 6-9 tissue samples from different parts of the species for the second and third generation transcription groups.
- If different individuals mix samples for genome assembly, the high heterozygosity may affect the size and effect of genome assembly.
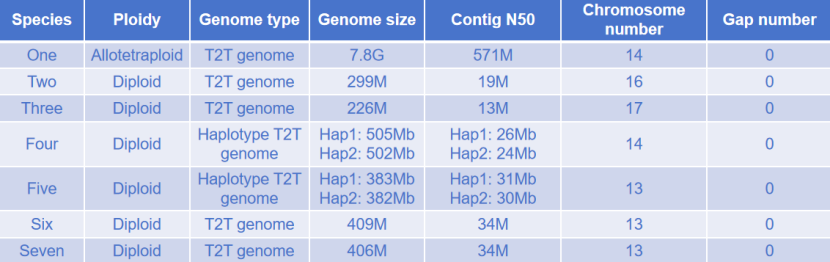 T2T genome project case
T2T genome project case
Three generations of DNA database sequencing products and genome De novo products have been deeply cultivated in the industry for many years. We take fast cycle, quality and quantity as our own responsibility, and serve customers and win-win cooperation as our purpose. We constantly hone our experimental skills, upgrade our computing power and improve the quality of analysis. We can provide detailed services for T2T genome detection, and you can contact us at any time.
References
- Bi Guiqi, Zhao Shijun, Yao Jiawei, Wang Huan and Dai Junbai. "Near telomere-to-telomere genome of the model plant Physcomitrium patens." Nature Plants (2024): 327-343. https://doi.org/10.1038/s41477-023-01614-7
- Chen Jia, Wang Zijian, Tang Kaiwei, Huang Wei and Lai Jinsheng. "A complete telomere-to-telomere assembly of the maize genome." Nature Genetics (2023) 1221-1231. https://doi.org/10.1038/s41588-023-01419-6
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.


 Summary of T2T genome in recent research
Summary of T2T genome in recent research
 T2T Assembly of Physcomitrium patens V6 Genome (Bi et al., 2023)
T2T Assembly of Physcomitrium patens V6 Genome (Bi et al., 2023)
 T2T assembly of Mo17 maize genome (Chen et al., 2023)
T2T assembly of Mo17 maize genome (Chen et al., 2023) Workflow of T2T genome analysis
Workflow of T2T genome analysis Recommended different T2T sequencing strategies
Recommended different T2T sequencing strategies T2T genome project case
T2T genome project case