In recent years, the second generation short reading long RNA sequencing (RNA-seq) technology has been widely used to study the dynamic changes, splicing trajectories and complex regulatory mechanisms of plant transcriptome. These studies emphasize the important role of plant transcriptome in responding to changes in development process and environmental conditions, and provide many important insights for understanding plant biology. However, due to the limitation of reading length and the bias that may be introduced in the process of reverse transcription of RNA into cDNA for library construction and amplification, it is more difficult to analyze co-transcription and post-transcription processing events.
Direct RNA Sequencing (DRS) is a long-reading and long-reading direct RNA sequencing technology based on Nanopore platform, which overcomes many limitations faced by traditional short-reading and long-reading RNA sequencing, such as reading length limitation and PCR amplification deviation. DRS can accurately detect the original full-length transcripts and analyze the complexity and diversity of plant transcriptome from many aspects (new isomers, poly A tail and RNA modification, etc.).
On August 18th, 2024, researchers from Guangxi University published a summary article entitled "Direct RNA Sequencing in Plants: Practical Applications and Future Perspectives" in Plant Communications. This paper systematically reviews the latest progress of DRS technology in analyzing the complexity and diversity of plant transcriptome, and puts forward a comprehensive workflow for processing plant DRS data. In addition, the application status of DRS technology in plant research and its possible future development direction are summarized.
Introduction of RNA Sequencing Technology
Short-reading and Long-reading RNA sequencing technology can analyze global RNA molecules and provide information on gene expression, alternative splicing and non-coding RNA functions. With the decrease of sequencing cost and the increase of sequencing depth, this kind of technology has become the mainstream tool of transcriptomics research in the past decade. However, due to the length of reading and the bias that may be introduced in the construction of cDNA library, the analysis of co-transcription and post-transcription processing events is limited. In contrast, the direct RNA sequencing (DRS) technology overcomes these limitations. DRS can sequence natural full-length RNA molecules, avoid the bias caused by PCR amplification, and identify RNA modifications by detecting current changes. DRS provides an effective method for studying gene expression patterns in different growth conditions or developmental stages, which is helpful for a deeper understanding of the function of plant genome, and improves the sensitivity and robustness of protein omics research of non-model plant species. In a word, DRS provides an unprecedented tool for describing many aspects of RNA biology, helps to reveal the relationship between genome and protein Group, and promotes the understanding of the diversity and complexity of plant transcriptome.
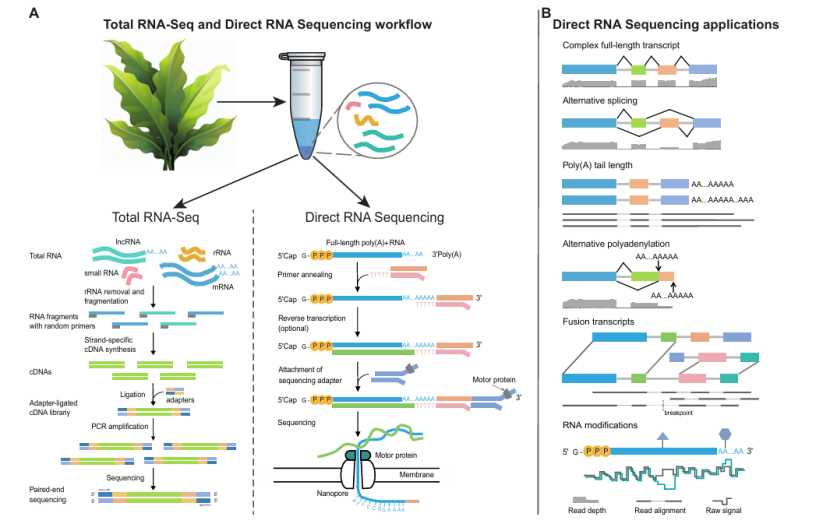 DRS method and its application in plants (Zhu et al., 2024)
DRS method and its application in plants (Zhu et al., 2024)
Service you may interested in
DRS Data Analysis Guide for Plants
DRS technology is widely used in transcriptome research of model and non-model plants, but there is a lack of comprehensive overview of plant DRS data processing flow at present. Therefore, this review proposes a three-stage data analysis framework: data preprocessing, comparison and biological significance research. First of all, the original DRS data is stored in Fast5 file, which can be converted into Fastq format by base recognition. Further comparison with reference genome or transcriptome is used to identify full-length transcripts and RNA modifications. These transcripts can be used for downstream analysis, such as quantifying the expression level, estimating the length of poly(A) and identifying modified bases, so as to explore the changes of RNA characteristics under different conditions.
For key data processing steps, it recommends specific tools. For example, Dorado is used for base recognition because of its high speed and advantages in methylation recognition. In correction, FMLRC2 is excellent in base-level accuracy, while isONcorrect is a tool for optimizing RNA data. In terms of quantification, RSEM shows the highest correlation, while NanoCount improves the accuracy of allele allocation. In addition, tools such as Tombo, CHEUI and TandemMod are used to find m5C modifications and predict m5C sites, and Nanom6A and m6Anet are used to identify m6A sites. Comparison tools such as minimap2 are commonly used, and Nanopolish is used to determine the length of poly(A) tail. The selection of these tools is based on performance benchmark test, frequency of use and practicality to assist further statistical analysis.
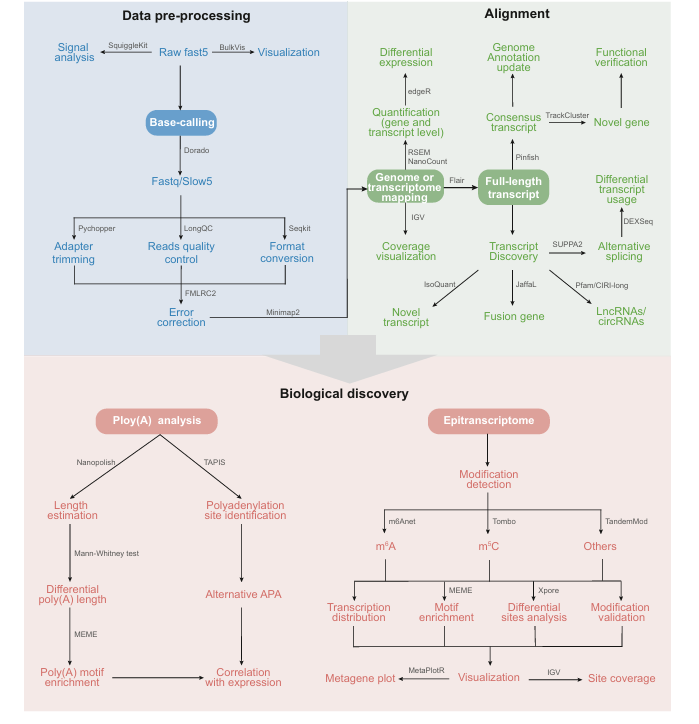 Systematic flow of plant DRS data analysis to obtain biological informaton (Zhu et al., 2024)
Systematic flow of plant DRS data analysis to obtain biological informaton (Zhu et al., 2024)
Full-length transcriptome is an ideal tool for gene model prediction and new gene identification, while direct RNA sequencing is especially suitable for discovering new complex isomers and analyzing the dynamic changes of transcriptome that are difficult to handle with short-read RNA-seq. Through DRS technology, researchers have found a large number of new transcripts in many plants, such as 22,360 in Phyllostachys pubescens, 16,432 in Platanus acerifolia, 578 in artichoke, 110,888 in strawberry and 38,500 in Arabidopsis thaliana. These new transcripts have significantly expanded the scope of the genome. In addition, DRS can also analyze the wide distribution of new transcripts produced by transposable elements (TEs) inserting introns in plants. The study of DRS in polyploid plants also shows obvious advantages, such as finding different alternative splicing events between hexaploid and decaploid in Platanus acerifolia.
DRS is also used to compare alternative splicing isomers between hybrids and their diploid parents, revealing important information related to the adaptive evolution of polyploid. In a word, DRS technology shows great ability in analyzing the complexity of plant transcriptome, which provides a unique perspective for understanding the biological role of full-length transcripts in plant development.
DRS Applications for Complex Plant Transcriptome
LncRNAs in plants participate in biological processes such as chromatin remodeling and transcription regulation through various mechanisms. DRS can identify new lncRNAs with complex structure and difficult to detect by short reading length technology. For example, 2,613 to 3,389 new lncRNAs were found in nine citrus species, and 796 new lncRNAs were identified in wheat, of which 29% contained TE insertions, which highlighted the ability of DRS to identify lncRNAs in polyploid and repetitive plant genomes. In addition, DRS is also used to detect circular RNA (circRNAs). Although there are technical challenges, these covalently closed circular structures can be effectively identified by special enrichment and reverse transcription methods, such as CIRI-long. For example, 470 circRNAs were identified in Phyllostachys pubescens, indicating that DRS has potential in identifying large circRNAs.
Identification of fusion transcripts
Fusion transcripts are hybrid molecules formed by the fusion of RNA products of two originally independent genomic regions or genes. These fusion events can be caused by genome rearrangement at DNA level, or by transcription splicing, transcription read-through and cis-splicing at RNA level. Compared with the short reading length method, DRS has a lower false positive rate in detecting fusion events and can detect full-length fusion transcripts. Although the research on the function of fusion transcripts in plants is limited at present, it has been revealed that they not only exist at the level of DNA and RNA, but also show low expression level and tissue specificity. For example, a new type of fusion transcript was identified in Arabidopsis thaliana by DRS technology, and it was found that transposable elements might promote the formation of fusion transcripts in grapes
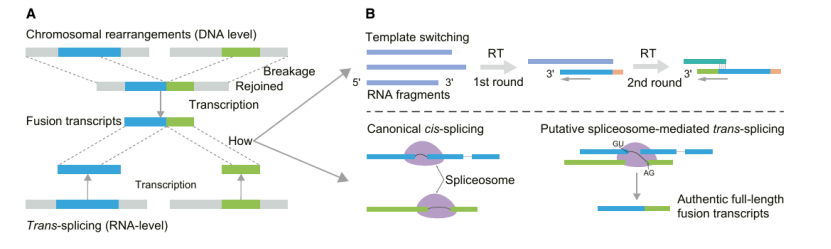 Types of fusion transcripts (Zhu et al., 2024)
Types of fusion transcripts (Zhu et al., 2024)
Functional characteristics of Poly (A) tail
Poly (A) tail is an important part of the 3' end of RNA molecule, and its length changes affect the output, stability and translation efficiency of RNA. DRS technology can capture full-length transcripts and reveal the changes of the tail length of poly (A) in different tissues. In addition, the tail length of poly (A) is gene-specific, and longevity mRNA usually has a short tail. Although the tail length distribution of different species is different, the tail length patterns of homologous genes are similar, indicating that poly (A) is conservative in the evolution process.
Variable polyadenylation (APA) increases the diversity of transcripts by changing the 3' splicing site, and has an important impact on the fate of RNA. DRS data revealed a large number of new APA events, such as the discovery of 109,880 APA loci in Arabidopsis thaliana. APA plays an important regulatory role in plant development and can respond to environmental signals, such as poplar tends to use remote APA sites under drought stress. Compared with traditional complex methods, DRS provides a simple and effective method for studying the dynamic changes of polyadenylation in plants.
Detection of RNA modification
DRS can quickly and accurately detect RNA modification without additional chemical treatment, which is a great progress compared with antibody-based technology. In eukaryotes, there are more than 170 reversible chemical modifications on RNA, which constitute the epigenome and participate in the regulation of gene expression. DRS can distinguish modified and unmodified bases, and detect many types of RNA modifications, including common m6A and m5C and other rare modifications, which affect RNA splicing, stability, movement and translation efficiency in plants. With the development of DRS technology, RNA modification can be detected efficiently, such as the recognition accuracy of m6A single base modification is 97%. In addition, DRS also revealed the relationship between modification and polyadenylation tail. For example, m6A-dependent poly (A) tail shortening will affect the stability of mRNA. In Arabidopsis thaliana and rice, DRS helps to analyze the functional mechanisms of m5C and m6A modifications under different conditions. For example, m5C plays an important role in long-distance mRNA movement, while m6A modification plays a key role in translation efficiency and abiotic stress response.
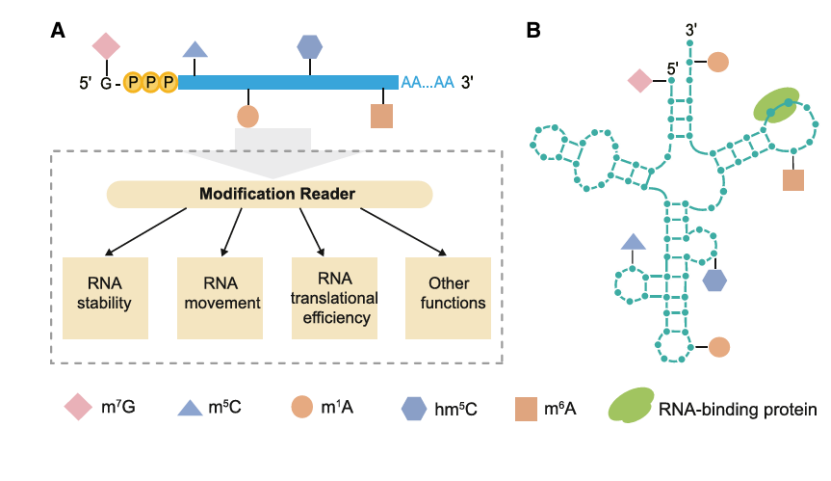 Various types of RNA modification (Zhu et al., 2024)
Various types of RNA modification (Zhu et al., 2024)
Challenges and Opportunities of DRS in Plants
The application of DRS in plant research is expected to bring revolutionary progress, especially in analyzing the complexity and diversity of transcriptome. However, this technology still faces some technical challenges, such as the need for a high amount of RNA (such as 300ng of poly(A) RNA). With the progress of technology, these limitations may be alleviated in the future. DRS can not only improve the classification, identification and expression quantification of microbial groups, but also discover new expression genes by detecting full-length reads, and explore the interaction between plants and their microbial groups, which is helpful to reveal the complex regulatory network of plant whole organisms.
DRS can also reduce the error sequences introduced by experiments in traditional methods, and show great potential in exploring the interaction between RNA modifications and other features (such as APA and secondary structure), and may reveal the role of these modifications in plant adaptive evolution. With the reduction of sequencing cost and the improvement of DRS resolution, it will be possible to construct pan-epigenetic transcriptome, which will help to understand the adaptability differences among different varieties. The application of single cell DRS will further enhance the understanding of plant cell heterogeneity, although the library construction scheme and data analysis algorithm still need to be optimized. In a word, DRS has great potential in plant research, and its future development will greatly promote our in-depth understanding of plant transcriptome and its function.
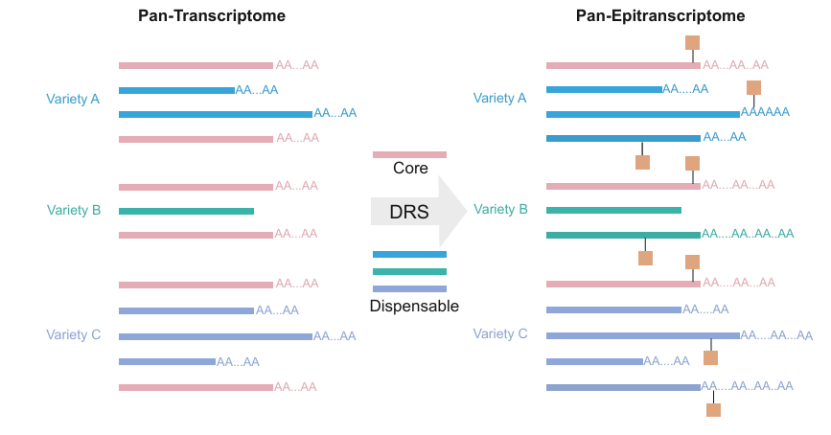 Illustrations of the pan-transcrip tome and pan-epitranscriptome (Zhu et al., 2024)
Illustrations of the pan-transcrip tome and pan-epitranscriptome (Zhu et al., 2024)
Conclusion
Although DRS technology is still in its infancy, it has shown its advantages over traditional RNA sequencing methods and great potential in plant transcriptomics research. It can not only help us understand the function of plant RNA metabolism more deeply, but also accelerate the development of crop varieties with strong adaptability and high economic benefits, and is expected to become an indispensable part of plant transcriptomics research. With the continuous development of this field, DRS will undoubtedly become a key force to promote the progress of plant science.
Reference
-
Zhu Xitong, Pablo Sanz-Jimenez., et al. "Direct RNA sequencing in plants: Practical applications and future perspectives." Plant communications 11 (2024): 101064. DOI: 10.1016/j.xplc.2024.101064
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.

 DRS method and its application in plants (Zhu et al., 2024)
DRS method and its application in plants (Zhu et al., 2024) Systematic flow of plant DRS data analysis to obtain biological informaton (Zhu et al., 2024)
Systematic flow of plant DRS data analysis to obtain biological informaton (Zhu et al., 2024) Types of fusion transcripts (Zhu et al., 2024)
Types of fusion transcripts (Zhu et al., 2024) Various types of RNA modification (Zhu et al., 2024)
Various types of RNA modification (Zhu et al., 2024) Illustrations of the pan-transcrip tome and pan-epitranscriptome (Zhu et al., 2024)
Illustrations of the pan-transcrip tome and pan-epitranscriptome (Zhu et al., 2024)