Since the Human Genome Project was launched in 1997, in less than 30 years of development, sequencing tools have been continuously upgraded. Taking human genome research as an example, the cost of sequencing has dropped significantly from several billion dollars to less than one hundred dollars. In less than 30 years, sequencing technology has experienced the development from the first generation of electrophoresis and capillary sequencing represented by Sanger sequencing, to the second generation of sequencing synthesis represented by illumina technology, and then to the third generation of sequencing technology, which provides the possibility for more species to splice perfect genomes.
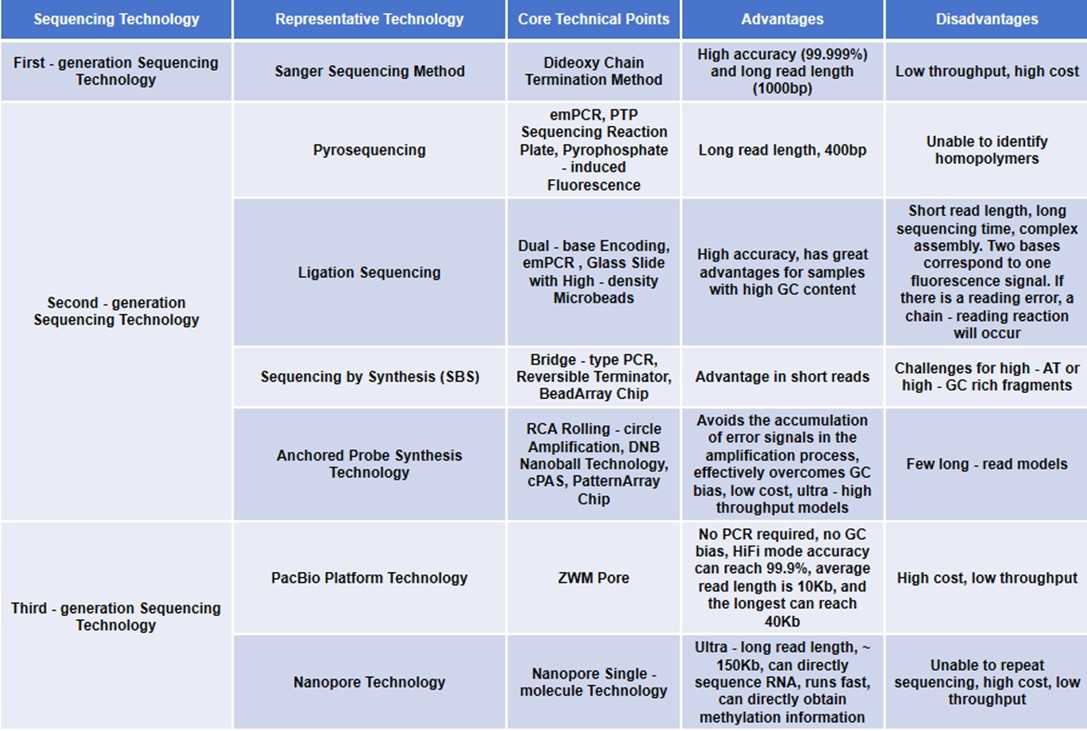 Summary of sequencing technology development.
Summary of sequencing technology development.
Service you may interested in
First Generation Sequencing Technology
Among the first generation sequencing, the most classic is Sanger sequencing, which was invented by Sanger in 1975 and used dideoxy chain termination method . Up to now, Sanger sequencing is still used to verify some genes, which is the "gold standard" of SNP detection and an irreplaceable, direct and accurate SNP detection method. Sanger is known as the father of sequencing, and his influence on biology is as great as that of Charles Darwin.
 Father of Sanger sequencing
Father of Sanger sequencing
Sanger sequencing is based on the method of double deoxynucleotide (ddNTP) termination. Specifically, dideoxynucleotide, T, G and C with different color labels were added into four separate reaction systems. Nucleotide extension reaction starts from a fixed point. If ddNTP is added in the extension process, because there is no 3'-OH (hydroxyl) on the base, the extension can't continue, thus randomly ending at a specific base, forming different series of nucleic acid fragments with a difference of one base. Then, these nucleic acid fragments with different lengths are separated by capillary electrophoresis, and the base sequence of the nucleic acid to be detected can be obtained by reading the colors of different base markers, and then the nucleotide sequence of the target region can be obtained.
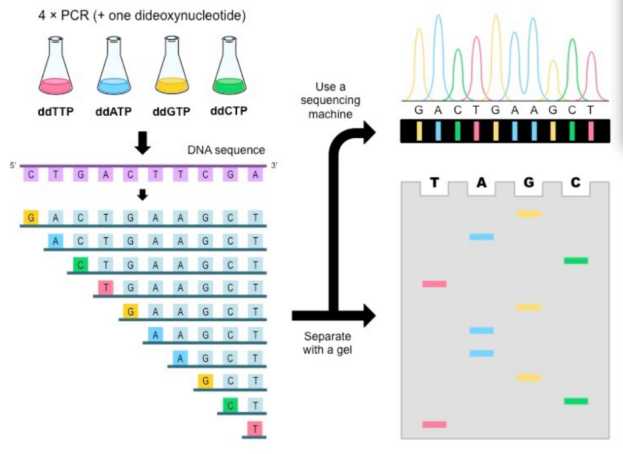 Sanger sequencing principle.
Sanger sequencing principle.
Although Sanger sequencing is particularly accurate, it is also costly. Therefore, for the whole genome sequencing, it is necessary to use the second-generation sequencing technology with low cost and Qualcomm.
Second Generation Sequencing Technology
In the second generation sequencing, 454 sequencing based on pyrophosphate sequencing technology and SOLID sequencing based on ligation sequencing have been basically stopped. The following mainly introduces illumina's sequencing while synthesizing and MGI's anchoring probe synthesis technology. The core of Illumina sequencing technology includes reversible blocking termination technology, sequencing while synthesizing and double-ended sequencing.
The sequencing principle is as follows: the genome of the sample is broken into an appropriate length, the end is repaired, the sequencing linker and index sequence are added to the fragment by PCR amplification, and then the NGS sequencing step is carried out, that is, sequencing while synthesizing. The specific process is to synthesize the first base, take photos, wash off impurities such as fluorescent substances, synthesize the second base, take photos, and repeat them continuously.
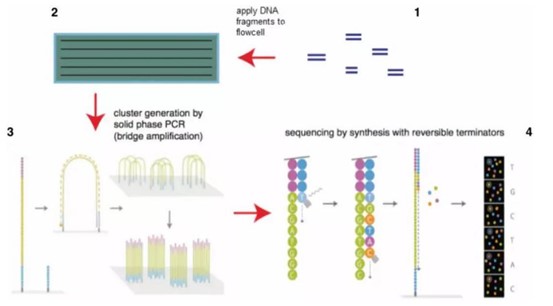 Schematic diagram of Illumina sequencing technology
Schematic diagram of Illumina sequencing technology
Third Generation Sequencing Technology
PacBio sequencing adopts single-molecule real-time (SMRT) sequencing technology. Its principle is that when the complex formed by DNA and polymerase is captured by zero-mode waveguides (ZMW), four different fluorescently labeled dNTP bases randomly enter the detection area through Brownian motion and combine with polymerase, and the fluorescent groups fall off during polymerization. Under the irradiation of laser, different colors are displayed, and the time for the base matching the template to form chemical bonds is much longer than that for other bases. Therefore, by counting the existence time of fluorescence signal, we can distinguish the matched base from the free base. The DNA template sequence can be determined by counting the relationship between four fluorescence signals and time.
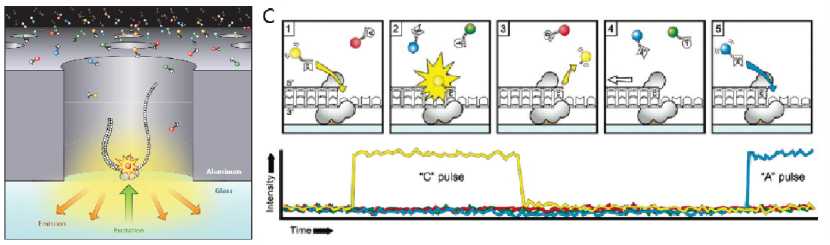 PacBio sequencing principle
PacBio sequencing principle
Due to the limitation of the reading length of DNA polymerase, the DNA template in the library with short fragment size can be tested many times to get multiple pass, which is called Circular Consensus Sequencing (CCS) mode. The DNA template in the library with long fragment size can only detect one pass, which is called continuous long read sequencing (CLR) mode.
PacBio HiFi (High Fidelity reads) is a sequencing method based on CCS mode introduced by PacBio 2019, which has both long reading length (10-20kb) and high accuracy (> 99% accuracy). In CCS sequencing mode, the restriction of enzyme reading length is far greater than the length of template DNA fragment, so polymerase will sequence around the template DNA in a rolling circle, and the inserted fragment will be sequenced many times. Random sequencing errors generated in a single sequencing, repeated reads generated by circular sequencing are self-correcting and corrected under the algorithm, and finally high-accuracy HiFi reads are obtained.
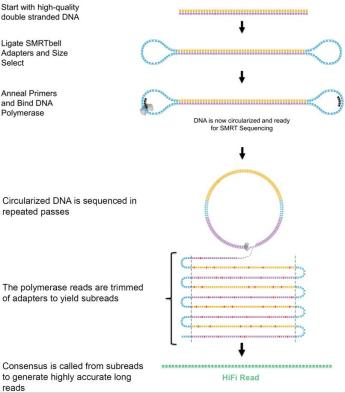 PacBio CCS sequencing mode
PacBio CCS sequencing mode
The principle of nanopore sequencing is that nanopore protein is used as a biosensor, and there are sensors on both sides of the nanopore to record voltage changes, which are inserted into the membrane formed by synthetic polymers. In addition, DNA nucleic acid molecules will be connected with Motor Protein. When voltage is applied, the motor protein will melt the double strands, so that the single strand of nucleic acid can pass through the specific nanopore protein under the action of electrophoresis. On the other hand, the moving speed of DNA/RNA molecules can be controlled, so that the bases can pass through the nanopore one by one, and a stable and reliable electrical signal can be generated. Because of the different charged properties of different bases, the sensor records these voltage changes, and by detecting the difference of electrical signals, the base types passing through the nanopore can be detected to realize sequencing.
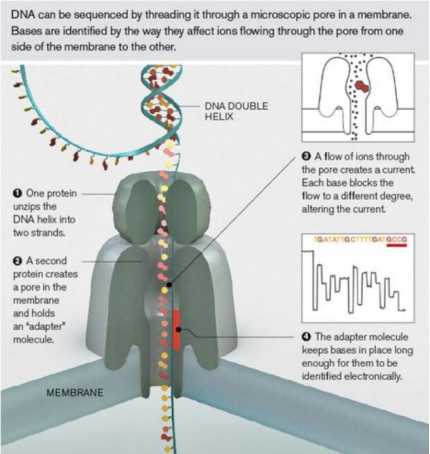 Principle of nanopore sequencing
Principle of nanopore sequencing
Compared with Pacbio, the biggest advantage of Nanopore technology lies in its reading length. In addition, it can directly sequence the original DNA and RNA without PCR amplification, thus avoiding amplification preference. The original base modification information is retained, and methylated cytosine can be directly read out.
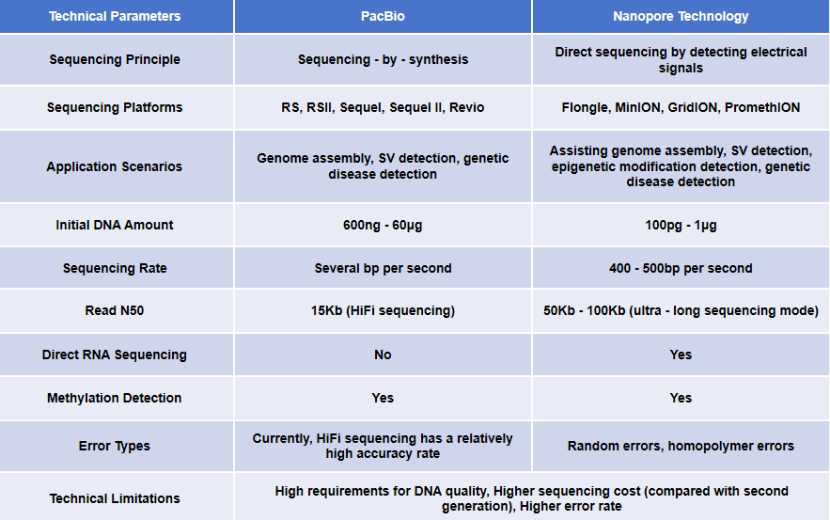 Summary of mainstream three-generation sequencing technology
Summary of mainstream three-generation sequencing technology
Agricultural Applications of Sequencing Technology
Promotes genome mapping of important agricultural species: Genomics research is a subject that studies how genes and genetic information in the genome of species are organically combined and how to determine their functions. This subject breaks the previous model of research at the level of a single gene and opens up a brand-new field aiming at the structure, expression and interaction of genome. With the emergence of this series of research achievements in animal and plant genomics, plant genomics research has also entered the post-genome era. People's cognitive level has also entered the dimensions of molecular structure, products and biochemical mechanisms from the original phenotypic description.
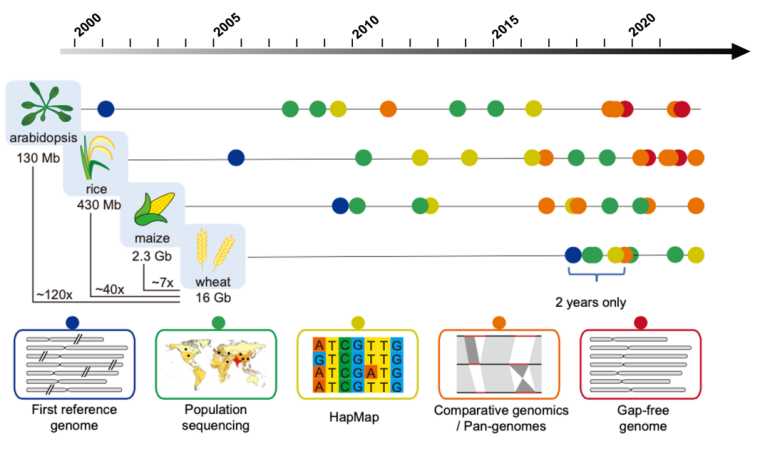 Progress timeline of genomics research of main plants
Progress timeline of genomics research of main plants
Accelerates the process of molecular breeding: Based on sequencing technology, animal and plant resequencing, transcriptome sequencing, comparative genome analysis and other technical means have been widely used in the research direction of molecular marker mining of important agronomic traits. Today, with the continuous reduction of sequencing cost, sequencing technology has been widely used in molecular breeding, improving breeding efficiency and accelerating breeding cycle.
The traditional breeding method is phenotypic breeding, which selects the best plant line according to the characters. The method is simple, but the breeding period is long and the accuracy is low. With the development of molecular biology, the identification of genes related to some important traits has promoted the rapid application of molecular marker-assisted selection. With the development of sequencing technology and the appearance of gene chips, scientists have developed the whole genome selection (GS) method, which is based on the whole genome molecular markers to help select the optimal strains or individuals with multiple traits. In addition, molecular breeding also includes transgenic, molecular design, gene editing and other technical means. Transgenic breeding is to introduce the desired gene into the receptor gene by some special means. Molecular design breeding is to find out the varieties that meet people's requirements through continuous practice, finally establish genes and put forward reasonable breeding methods.
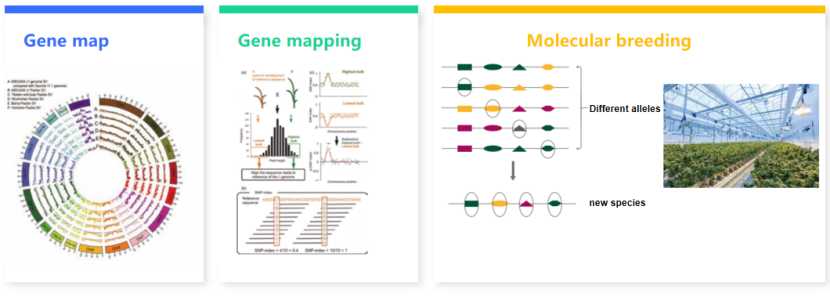 Sequencing technology runs through all major links of gene-assisted breeding
Sequencing technology runs through all major links of gene-assisted breeding
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.


 Summary of sequencing technology development.
Summary of sequencing technology development. Father of Sanger sequencing
Father of Sanger sequencing Sanger sequencing principle.
Sanger sequencing principle. Schematic diagram of Illumina sequencing technology
Schematic diagram of Illumina sequencing technology PacBio sequencing principle
PacBio sequencing principle PacBio CCS sequencing mode
PacBio CCS sequencing mode Principle of nanopore sequencing
Principle of nanopore sequencing Summary of mainstream three-generation sequencing technology
Summary of mainstream three-generation sequencing technology Progress timeline of genomics research of main plants
Progress timeline of genomics research of main plants Sequencing technology runs through all major links of gene-assisted breeding
Sequencing technology runs through all major links of gene-assisted breeding