Introduction to Antibody Screening Sequencing or Phage Display Library Screening
Phage display is a popular and effective laboratory tool for discovering peptides that bind to any target. This method is typically based on phage particle libraries containing millions or even billions of exogenous peptides bonded to phage surface proteins. The highly diverse library is shortened to a few leads through several rounds of selection, also known as biopanning, and amplification is required to enhance phage clones that present target-binding peptides, in order to assess the binders with high affinity and specificity. Given that antibody fragments such as scFv or Fab are quickly expressed and displayed on phage, phage display technology plays a critical role in the discovery and engineering of antibodies.
The analysis of peptide sequences in the library is crucial to phage display. Individual phage clones must be isolated for conventional Sanger sequencing. It's a time-consuming, small-scale process that relies on repeated amplification. The method's limitations include the generation of unintended selective bias against certain antibody sequences in bacteria and the inability to evaluate more than a hundred library clones.
When it comes to analyzing phage-display screens, Illumina deep-sequencing has a lot of benefits, especially for large sequence collections For starters, it allows for phage display experiments with fewer, and eventually just one, selection rounds, potentially avoiding problems caused by unwanted bias and collapse of diversity during the amplification process, allowing for the discovery of ligands that were previously lost or under-represented in phage display screens. Furthermore, NGS platforms can characterize up to 106–108 sequences in a single run, allowing for a much more representative, and thus useful, content coverage.
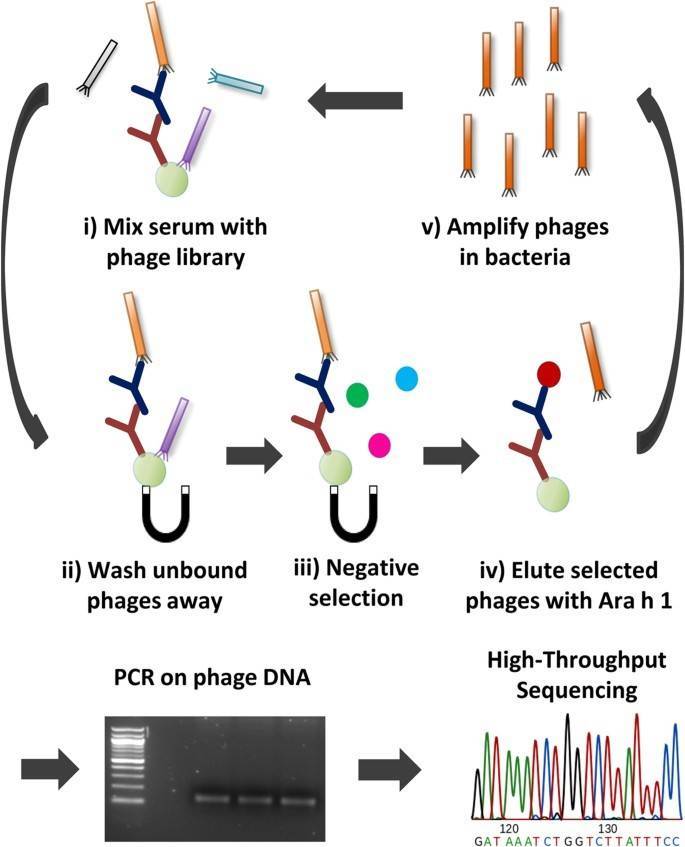
Figure 1. An overview of the phage selection process. (Christiansen, 2015)
General Workflow for Antibody Screening Sequencing
The basic guidelines for antibody screening sequencing are as follows:
- Utilizing various trypsins, the antibody protein is first digested into overlapping peptides.
- LC-MS/MS digestion, followed by fragmentation optimization
- De novo peptide sequencing is used to identify overlapping peptides.
- Automated antibody sequence assembly using peptides that have been categorized
- Data is analyzed using cutting-edge computational algorithms to quickly determine the amino acid sequence of the digested peptide. Applications of Antibody Screening Sequencing
Antibody discovery by sequencing immune repertoires
Various immune compartments from distinct mammalian hosts have been probed post-immunization in order to find new monoclonal antibodies using high-throughput sequencing and bioinformatics assessment. Three different methods have been proven to be effective in this regard.
Sequencing recombinant antibody repertoires
Ig-seq can be employed to investigate a variety of recombinant antibody libraries, including naive, focused, immune, semi-synthetic, and fully synthetic libraries, in addition, to directly mining natural B-cell-derived antibody repertoires of animals and humans. In vitro screening platforms such as phage, yeast, and ribosome display are suitable with these repertoires. One of the main goals of these sequencing efforts is to capture and deduce library-quality metrics that indicate the likelihood of effectively isolating antibodies with preferred characteristics from a provided recombinant library.
Combining high-throughput screening and sequencing for monoclonal antibody discovery and engineering
For high-throughput sequencing to offer a meaningful in-depth perspective of possible antigen-specific library varieties, diversity must be considerably lowered in order to obtain the full ability of a given natural or recombinant antibody library (usually in the range of 106–1011 clones). As a result, integrating Ig-seq with a phenotype-based sequence filter and selection pressure, such as those supplied by phage or yeast display, is a feasible method for selectively interrogating antibody libraries in depth.
References:
- Fischer N. Sequencing antibody repertoires: the next generation. InMAbs 2011 Jan 1 (Vol. 3, No. 1, pp. 17-20). Taylor & Francis.
- Parola C, Neumeier D, Reddy ST. Integrating high‐throughput screening and sequencing for monoclonal antibody discovery and engineering. Immunology. 2018 Jan;153(1).
- Thompson KL, Rosenzweig BA, Pine PS, et al. Use of a mixed tissue RNA design for performance assessments on multiple microarray formats. Nucleic acids research. 2005 Jan 1;33(22).
- Christiansen A, Kringelum JV, Hansen CS, et al. High-throughput sequencing enhanced phage display enables the identification of patient-specific epitope motifs in serum. Scientific reports. 2015 Aug 6;5(1).
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.


 Sample Submission Guidelines
Sample Submission Guidelines

