Fluorescent Quantitative PCR (qPCR) technology allows precise tracking of fluorescence signal changes during the PCR process in real-time. In the context of fluorescent quantitative PCR analysis, three key types of curves are pivotal: the amplification curve, the melt curve, and the standard curve.
The Basics of qPCR Amplification
The qPCR amplification process, especially when using SYBR Green I dye, has two main stages: amplification and melting.
Amplification Phase
The amplification phase uses a two-step method. At the end of each cycle, fluorescence signals are detected and recorded to form an amplification curve.
Melting Phase
Upon completion of the amplification phase, a separate melting reaction program is initiated. This phase monitors the fluorescence signal changes during the denaturation process (the step where the DNA strands separate) of the double-stranded DNA products, leading to the generation of a melt curve.
Amplification Curve
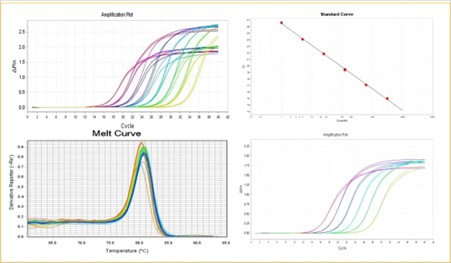
The amplification curve provides a comprehensive visualization of the dynamic progress within the polymerase chain reaction (PCR). This curve is graphically represented with the number of cycles on the x-axis and the corresponding real-time fluorescence signal intensity on the y-axis.
In an ideal theoretical framework, reaction products in PCR experience exponential amplification. However, as the number of cycles increases, several factors—such as the inactivation of DNA polymerase, the depletion of deoxynucleotide triphosphates (dNTPs) and primers, and the accumulation of inhibitory by-products—adversely affect amplification efficiency. Consequently, the pace of product generation diminishes, culminating in an S-shaped amplification curve.
The amplification curve is divided into four distinct phases:
Baseline Phase: This initial phase corresponds to the flat portion of the curve. Here, the fluorescence signal from the amplification process is masked by background fluorescence, rendering any changes in product quantity undetectable.
Exponential Phase: During this phase, PCR enters a stage of exponential amplification, and the curve begins its ascent. The quantity of product generated in each cycle is directly proportional to the initial amount of template, adhering to an exponential relationship. Notably, a linear relationship is evident between the logarithmic values of the PCR product and the initial template concentration.
Linear Phase: In the later stages of PCR, the reaction efficiency declines due to the depletion of reaction components and the inhibition by accumulated products. Consequently, product accumulation deviates from the exponential growth pattern, resulting in a decoupling of the amount of PCR product from the initial template quantity.
Plateau Phase: During this final phase, the PCR reaction reaches cessation, with no additional increase in product regardless of further cycling. The onset of the plateau phase and the final plateau level are influenced by a myriad of factors intrinsic to the PCR system, contributing to variability across different reactions.
Key Terms in the Amplification Curve
To understand the amplification curve, you need to know a few key terms.
Baseline
The baseline refers to the region in the amplification curve where the fluorescence signal remains relatively stable due to the background fluorescence. This typically occurs during the initial 3 to 15 cycles of PCR, where changes in fluorescence are minimal and the curve appears nearly linear. The baseline can either be automatically generated or manually set. When manually adjusting, it is crucial to ensure that the baseline is configured to eliminate background fluorescence without masking the amplification signals of non-fluorescent products. For reactions involving multiple genes, the baseline should be set separately for each gene. Modern versions of quantitative PCR software allow for automatic optimization of baseline settings for individual samples.
Threshold
The threshold is defined as a fluorescence detection boundary set at an appropriate point within the exponential phase of the amplification curve. It can be automatically or manually set by the instrument. The threshold is usually set to 10 times the standard deviation of the baseline fluorescence signal. For reactions targeting different genes within the same experiment, individual thresholds may be set, but the same threshold must be applied to the amplification of a particular gene across all samples.
Ct Value (Cycle Threshold Value)
The Ct value shows how many cycles it takes for the fluorescence signal to reach the threshold. The Ct value typically falls within the early stages of the exponential phase, where small variations between samples have not yet amplified and the amplification efficiency remains relatively constant. A linear relationship exists between the logarithm of the starting template copy number and the Ct value. Ct values are consistent, and the typical range for analysis is between 15 and 35 cycles. Extremely high or low Ct values may compromise the accuracy of quantitative results.
Characteristics of a Good Amplification Curve
An ideal amplification curve should display:
Flat Baseline Period: The baseline should be flat or slightly declining, without a significant upward trend.
Clear Exponential Phase: The exponential phase should exhibit a sharp rise, with a steep gradient and a clearly defined inflection point.
Smooth Overall Curve: The linear phase should gradually level off, and the plateau phase should be nearly flat.
Good Reproducibility: Replicate wells should show consistent amplification curves during the exponential phase, with minimal variation in Ct values (preferably no more than a 0.5 Ct difference).
Interpreting the Melt Curve
In accordance with established primer design principles, the length of the amplification product in fluorescent quantitative PCR (qPCR) generally spans between 80 and 200 base pairs (bp). Correspondingly, the melting temperature ™ of these products typically falls within the range of 80 to 90°C. If no amplification is seen in the negative control, the melt curve helps verify the specificity of the amplified product.
Single Peak: A melt curve displaying a single, distinct peak between 80 and 90°C signifies robust specificity of the PCR product, indicating successful amplification and alignment with the intended target sequence.
Double Peak (Main Peak at 80–90°C, Secondary Peak Below 80°C): The occurrence of a primary peak within the 80 to 90°C range, accompanied by an additional secondary peak below 80°C (commonly between 60 to 75°C), suggests the formation of primer-dimers. To address this issue, optimization strategies such as increasing the annealing temperature, reducing primer concentration, or redesigning the primers may be employed.
Double Peak (Main Peak at 80–90°C, Secondary Peak Above 90°C): The presence of a secondary peak above 90°C on the melt curve is indicative of non-specific amplification, potentially arising from genomic DNA contamination within the template. To mitigate such non-specific amplification, it is advisable to employ strategies such as designing primers that span intronic regions or removing genomic DNA from the sample.
Standard Curve
A standard curve is typically generated by serially diluting a standard sample to at least five different concentrations. The starting copy number is plotted on the x-axis, while the corresponding Ct values are plotted on the y-axis. This results in the construction of a standard curve that establishes a linear relationship between Ct values and the initial copy number of the target sequence.
Several parameters of the standard curve can be utilized to assess the performance of a fluorescent quantitative PCR (qPCR) system.
Slope
The slope of the standard curve represents the amplification efficiency of the qPCR system. The theoretical value for a perfect amplification efficiency is -3.32, which corresponds to an amplification efficiency of 100%. This indicates that the amount of PCR product doubles with each cycle.
Amplification Efficiency Below 100%: When the amplification efficiency is below 100%, this may indicate suboptimal PCR performance, which could be due to factors such as poorly designed primers, unsuitable reagents, or inappropriate reaction conditions. Additionally, inaccuracies in the dilution of the standard sample may also contribute to this issue.
Amplification Efficiency Above 100%: If the amplification efficiency exceeds 100%, it suggests the presence of inhibitory factors in the reaction system. These factors may include poor template quality, excessive template concentration, or the accumulation of reaction inhibitors. In some cases, non-specific amplification may also lead to an overestimation of the amplification efficiency, which can be further investigated using the melt curve analysis.
It is important to note that not all template molecules will undergo perfect doubling during each cycle. Generally, an amplification efficiency between 90% and 110% is considered acceptable for data analysis.
qPCR Services for Microbial Functional Genes
CD Genomics offers comprehensive and scientifically rigorous services in the field of microbial functional gene quantification through qPCR. With years of experience and technical expertise in microbial genomics, CD Genomics provides robust technical support and ensures high-quality results.
Clients need only to provide tissue samples along with relevant gene information. CD Genomics will conduct all experimental procedures and analyses, delivering a detailed experimental report upon completion of the analysis.
Conclusion
Understanding the key components of qPCR, such as the amplification curve, melt curve, and standard curve, is essential for accurate and reliable DNA quantification. With proper interpretation of these curves, researchers can ensure the specificity and efficiency of their PCR reactions, leading to more accurate and reproducible results. Whether you're working in microbial genomics or other fields, qPCR remains a crucial tool for DNA analysis.


 Sample Submission Guidelines
Sample Submission Guidelines

