Illumina's Next-Generation Sequencing (NGS) workflow is a sophisticated and highly efficient process that enables researchers to unravel the mysteries of genetics with speed and accuracy. The workflow on Illumina platforms can be broken down into three main steps: library preparation, sequencing, and data analysis.
Principles of Illumina Sequencing Technology
The fundamental principle of Illumina sequencing technology revolves around the utilization of fluorescently labeled nucleotides that possess reversible terminators. This approach shares the core concept of "sequencing by synthesis-as-you-go," akin to the Sanger method. However, unlike Sanger, this technique involves temporary termination of DNA strand extension after incorporating each uniquely modified nucleotide. Once the added nucleotides are optically detected using specific fluorescent markers, the terminator molecules are cleaved, allowing the synthesis of the new strand to resume for the subsequent round of nucleotide addition.
To simultaneously detect nucleotide incorporation in millions of sequencing reactions, dATP, dCTP, dGTP, and dTTP are each tagged with distinct fluorescent labels, enabling the differentiation of nucleotides based on the emitted fluorescent signals. These fluorescent labels, along with the reversible terminator molecules, are bound to the nucleotides through the same chemical bonds. As a result, following the integration and detection of each nucleotide during the sequencing cycle, both the fluorescent labels and terminator molecules can be cleaved off simultaneously in a single reaction, preparing the way for the integration of the next nucleotide.
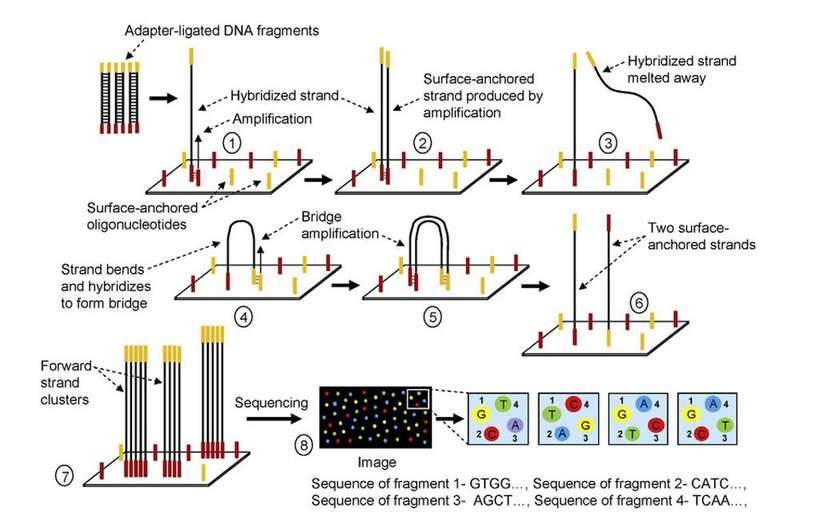 3 Principles of Illumina Solexa sequencing. (Choudhuri Supratim, 2014)
3 Principles of Illumina Solexa sequencing. (Choudhuri Supratim, 2014)
How Does Illumina Sequencing Work?
Sequencing reactions on the Illumina NGS System occur within a flow cell. The flow cell contains microfluidic channels, often referred to as lanes, where the sequencing reaction takes place and sequencing signals are collected through scanning.
Within these channels, the upper and lower surfaces are covered with a "lawn" of oligonucleotide sequences, which complement the anchoring sequence components at the junction. When the sequencing library is introduced into each channel, the DNA templates within the library bind to these oligonucleotide sequences, becoming immobilized on the channel surface.
Following immobilization, each DNA template molecule undergoes clonal amplification via a process called "bridge amplification." This process generates up to 1,000 identical copies of the template in close proximity, forming clusters of less than 1 micron in diameter. These clusters serve as the fundamental detection units during the sequencing process, providing sufficient signal strength for base recognition.
Library Preparation
The first step in the Illumina NGS workflow is library preparation, a crucial stage that ensures the DNA or RNA samples are compatible with the sequencer. This process involves fragmenting the DNA into smaller pieces, followed by the addition of specific adapters to the ends, creating the sequencing library.
In Illumina sequencing, the adapters contain complementary sequences that allow the DNA fragments to bind to the flow cell, which is where the sequencing process takes place. Once the fragments are bound, they undergo amplification and purification. To optimize resources, multiple libraries can be mixed together and sequenced in the same run, a process called multiplex analysis. Unique dual indexes (UDI) are added to each library during junction ligation, serving as barcodes to distinguish the different libraries during data analysis.
UDI is particularly useful in multiplex analysis to reduce sample mismatches due to label jumping, especially in instruments with patterned flow cells like the NovaSeq 6000 system. Additionally, the inclusion of molecular barcodes to each molecule in the library enhances the sensitivity of variant detection and helps eliminate PCR duplicates and low-frequency variants.
Sequencing
During the sequencing step of the NGS workflow, the prepared library is up-sampled into a flow cell and placed inside the sequencer. The cluster generation process amplifies clusters of DNA fragments, producing millions of single-stranded DNA copies. Most Illumina sequencing instruments can perform cluster generation automatically.
Sequencing-by-synthesis (SBS) is the method used during the actual sequencing process. Chemically modified nucleotides bind to the DNA template strand through natural complementarity. Each nucleotide has a fluorescent marker and a reversible terminator, preventing the incorporation of the next base. The fluorescent signal indicates the type of nucleotide added, and the terminator is then cleaved, allowing the next base to bind.
After reading the forward DNA strand, the read is washed away, and the process is repeated to read the reverse strand, making it a double-ended sequencing method.
Data Analysis
At the completion of sequencing, the instrument software performs base detection, identifying the nucleotides present (known as primary analysis) and predicting the accuracy of this base detection. The generated sequencing data can then be imported into standard analysis tools for further processing or custom analysis pipelines can be created (known as secondary analysis). Researchers often utilize intuitive data analysis applications (tertiary analysis) to interpret and extract meaningful insights from the NGS data. Data analysis is a critical phase as it allows researchers to identify genetic variations, mutations, and structural rearrangements within the genome, leading to important discoveries in fields such as disease genomics, personalized medicine, and agriculture.
Reference:
-
Choudhuri, Supratim. Bioinformatics for beginners: genes, genomes, molecular evolution, databases and analytical tools. Elsevier, 2014.
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.


 Sample Submission Guidelines
Sample Submission Guidelines
 3 Principles of Illumina Solexa sequencing. (Choudhuri Supratim, 2014)
3 Principles of Illumina Solexa sequencing. (Choudhuri Supratim, 2014)