Differential gene expression analysis is a fundamental technique in bioinformatics that plays a crucial role in deciphering the complex mechanisms underlying various biological processes. By comparing gene expression levels between different conditions or treatments, researchers can identify differentially expressed genes, shedding light on key biological pathways and providing valuable insights into normal physiology and disease states. In this article, we will explore the principles, methods, and applications of differential gene expression analysis, highlighting its significance in advancing our understanding of the intricate molecular mechanisms of life.
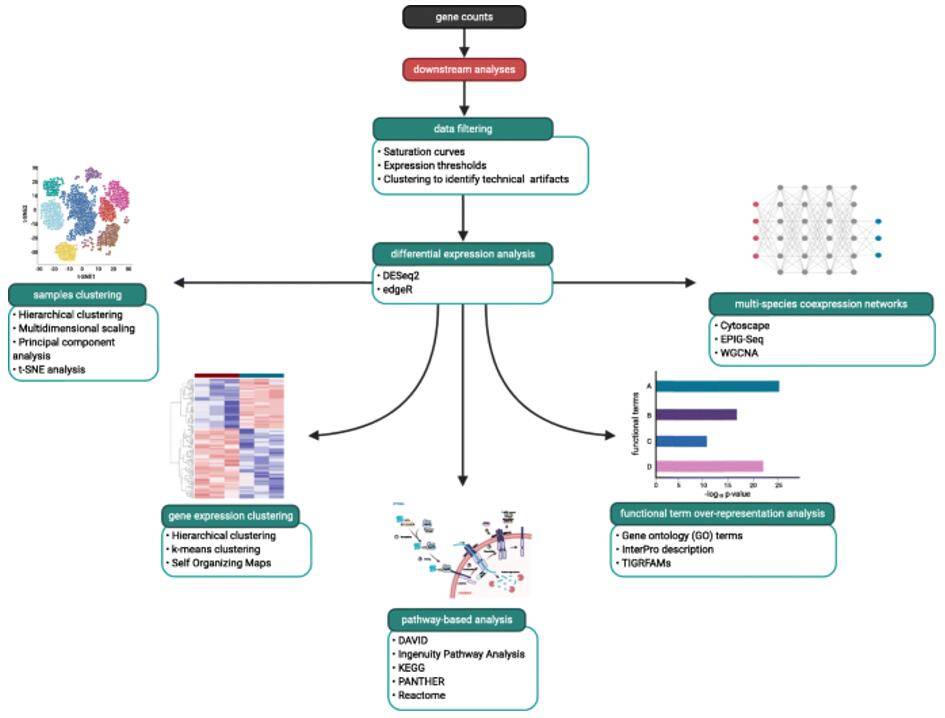 A general workflow showing examples of downstream analyses for a typical multi-species RNA-Seq analysis. (Chung et al., 2021)
A general workflow showing examples of downstream analyses for a typical multi-species RNA-Seq analysis. (Chung et al., 2021)
What is DGE Analysis?
Gene expression refers to the process by which information encoded in DNA is converted into functional proteins or non-coding RNAs. Differential gene expression analysis aims to identify genes that exhibit significant changes in expression levels between two or more conditions, such as different cell types, disease states, or experimental treatments. These genes are crucial in unraveling the biological factors driving phenotypic differences and can serve as potential biomarkers or therapeutic targets.
Methods for Differential Gene Expression Analysis
Microarray Analysis
Microarrays have been widely used for gene expression profiling. By hybridizing labeled cDNA or RNA samples onto a microarray chip containing thousands of gene probes, researchers can measure the relative abundance of transcripts. Statistical algorithms are employed to identify differentially expressed genes.
RNA Sequencing (RNA-Seq)
RNA-Seq utilizes next-generation sequencing (NGS) technologies to sequence the cDNA generated from the extracted RNA. The RNA is fragmented, and cDNA libraries are prepared by reverse transcription, adapter ligation, and PCR amplification. The resulting libraries are then sequenced on platforms like Illumina, Ion Torrent, or Pacific Biosciences.
Please refer to our article RNA-Seq for Differential Gene Expression Analysis: Introduction, Protocol, and Bioinformatics for more information.
CD Genomics offers Differential Gene Expression (DGE) Analysis service, leveraging data from both sequencing technologies and Microarray experiments. This integrated service enables a comprehensive examination of gene expression patterns, shedding light on the intricate molecular dynamics underlying diverse biological conditions.
Statistical Analysis of Differential Gene Expression
Statistical methods play a critical role in identifying genes that exhibit significant expression differences. Techniques such as t-tests, analysis of variance (ANOVA), and non-parametric tests are commonly employed to assess the statistical significance of gene expression changes. Multiple testing corrections help control false discovery rates and improve the reliability of results.
Here are some commonly used methods for differential expression analysis:
EdgeR
EdgeR is a widely used method based on the negative binomial (NB) distribution. It uses generalized linear models (GLMs) to model the count data obtained from RNA-Seq experiments. EdgeR is particularly useful for experiments with a small number of biological replicates and is known for its robustness in handling both common and rare genes.
DESeq2
DESeq2 is another popular method that utilizes the negative binomial distribution. It employs shrinkage estimators to improve the estimation of dispersion, especially for genes with low counts. DESeq2 uses a Wald test or likelihood ratio test to assess differential expression. It is known for its ability to handle experiments with a larger number of replicates and provides accurate results even in the presence of batch effects.
limma-voom
limma-voom is a differential expression analysis method that combines linear modeling and empirical Bayes methods. It incorporates a precision weighting approach (voom) to transform count data into log-transformed, normalized data suitable for linear modeling. limma-voom is particularly effective when dealing with experiments with a limited number of replicates and has been widely used in both microarray and RNA-Seq analyses.
baySeq
baySeq is a Bayesian approach for differential expression analysis. It assumes a negative binomial model and estimates posterior probabilities of differential expression. baySeq is useful for detecting differential expression when dealing with low replicate numbers or high levels of biological variability. It provides posterior probabilities for each gene, allowing researchers to make informed decisions based on statistical evidence.
EBSeq
EBSeq is another Bayesian approach that models differential expression using the negative binomial distribution. It utilizes empirical Bayes estimation and provides a posterior probability for each gene being differentially expressed. EBSeq is particularly useful when dealing with small sample sizes and exhibits good performance in identifying differentially expressed genes.
Applications of Differential Gene Expression Analysis:
Disease Research
Differential gene expression analysis provides crucial insights into the molecular mechanisms underlying various diseases. By comparing gene expression profiles between healthy and diseased tissues, researchers can identify dysregulated genes and pathways associated with pathogenesis. These findings can lead to the development of novel diagnostic markers and therapeutic targets.
Drug Discovery
Identifying differentially expressed genes in response to drug treatments can reveal the molecular targets and mechanisms of action. This knowledge aids in drug discovery and the development of personalized medicine approaches.
Developmental Biology
Differential gene expression analysis enables the exploration of gene regulatory networks during embryonic development, tissue differentiation, and organogenesis. It helps uncover key genes and signaling pathways involved in these processes.
Environmental Studies
By comparing gene expression patterns in response to environmental factors, researchers can gain insights into the impact of pollutants, toxins, and stressors on living organisms.
Challenges and Future Directions
Despite its tremendous potential, analyzing differential gene expression poses various challenges, including data normalization, batch effects, and the need for robust statistical approaches. Integrating multi-omics data and developing sophisticated computational tools will further enhance the accuracy and biological interpretation of differential gene expression analysis.
Reference:
- Chung, Matthew, et al. "Best practices on the differential expression analysis of multi-species RNA-seq." Genome biology 22.1 (2021): 1-23.
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.


 Sample Submission Guidelines
Sample Submission Guidelines
 A general workflow showing examples of downstream analyses for a typical multi-species RNA-Seq analysis. (Chung et al., 2021)
A general workflow showing examples of downstream analyses for a typical multi-species RNA-Seq analysis. (Chung et al., 2021)