CRISPR Gene Editing Technology
In recent years, the CRISPR technology has emerged as a prominent innovation in the field of biotechnology. This technique utilizes guide RNA (gRNA) to direct Cas nucleases with precision, enabling specific DNA modifications of target genes. The CRISPR-Cas system is underpinned by two core components: gRNA and CRISPR-associated Cas nucleases.
The gRNA is responsible for identifying specific regions of the target DNA and directing the Cas nuclease to the appropriate loci for editing. The structure of gRNA is comprised of two segments: CRISPR RNA (crRNA), a 17-20 nucleotide sequence highly complementary to the target DNA; and trans-activating CRISPR RNA (tracrRNA), which serves as a binding scaffold for the Cas nuclease, ensuring its precise localization.
The binding process between the gRNA and the target sequence relies upon the presence of a protospacer adjacent motif (PAM) in the genomic DNA. Upon successful binding, the Cas9 nuclease induces double-strand breaks (DSBs) within the DNA. In the presence of an appropriate DNA template, these DSBs can engage endogenous DNA repair mechanisms, potentially resulting in gene knockouts or facilitating precise gene modifications, such as frameshift mutations or sequence insertions.
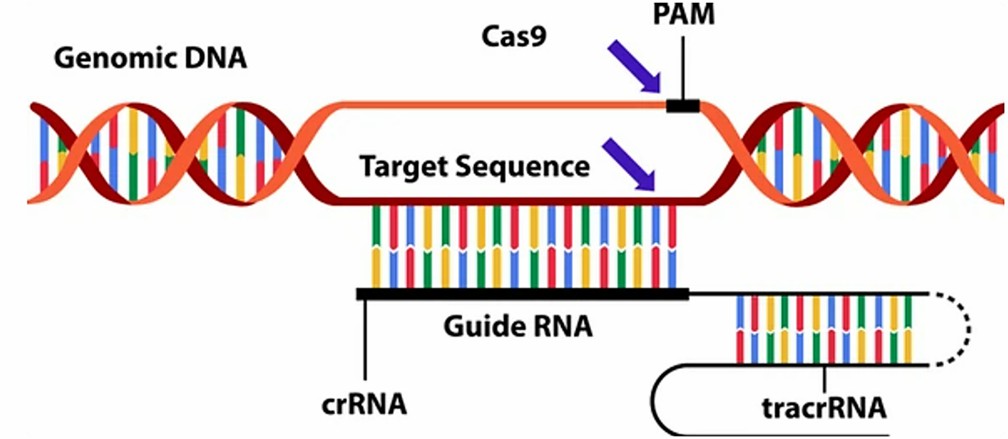 Figure 1. CRISPR gene editing technology
Figure 1. CRISPR gene editing technology
CRISPR Library Screening Technology
CRISPR library screening represents a significant application of the CRISPR/Cas9 system, demonstrating its unique utility in the field of gene editing. In CRISPR experiments, the crRNA portion of the gRNA serves as a customizable component, conferring the specificity required for the experiment. Researchers have successfully utilized CRISPR/Cas9 technology to construct whole-genome knockout libraries or gene knockout libraries closely associated with specific functions in the target species.
Subsequently, through functional screening and enrichment procedures, followed by PCR amplification and deep sequencing analysis, it becomes feasible to accurately identify genes associated with specific phenotypes. This comprehensive process is referred to as sgRNA library screening.
The application of high-throughput CRISPR screening has emerged as a pivotal technique within the genomic editing domain. This method utilizes high-throughput synthesized sgRNA libraries, which are efficiently delivered into cells via lentiviral vectors. Following transduction, cells undergo various selective pressures, such as drug selection and fluorescence-activated cell sorting (FACS), to enrich for target genes. Post-selection, cell samples are collected and subjected to next-generation sequencing (NGS) to analyze the differential enrichment or depletion of sgRNAs between experimental and control groups. This process elucidates host factors closely associated with the selective conditions.
The implementation of this methodology has significantly enhanced the precision and efficiency of screening clones while reducing experimental costs and time investment. The significance of CRISPR library screening lies in its capacity to rapidly identify target genes and investigate their functions, thereby providing robust support for research in gene editing and therapeutic interventions.
Applications of CRISPR sgRNA Library Screening
Exploration of Drug Mechanisms of Action: Drug Target Identification and Validation
The employment of CRISPR sgRNA library screening is pivotal in elucidating the molecular targets of pharmacological agents. Through the induction of precise gene knockouts, researchers can observe resultant phenotypic alterations, which shed light on the interactions between drugs and their respective molecular targets. This methodology serves to validate both the efficacy and specificity of therapeutic compounds, thereby enhancing the precision of drug development processes.
Identification of Cancer Therapy Targets: Analysis of Upstream and Downstream Regulatory Mechanisms
Within the realm of oncology research, CRISPR sgRNA libraries are harnessed to identify genes implicated in cancer progression and resistance mechanisms. This technique facilitates the dissection of intricate regulatory pathways, thereby providing insights into both upstream and downstream effectors that could constitute potential therapeutic targets. Such insights are invaluable for the development of more effective cancer treatments.
Investigation of Metabolic Disease Treatment Mechanisms: Analysis of Metabolic Pathway Regulation
The application of CRISPR sgRNA library screening in the context of metabolic disease research enables the identification of critical regulatory elements within metabolic pathways. By systematically knocking out specific genes, researchers can elucidate the roles of individual genetic components in the regulation of metabolism. This approach is instrumental in identifying potential targets for therapeutic intervention, thereby advancing the understanding and treatment of metabolic disorders.
CD Genomics High-Throughput sgRNA Library Screening Service
CD Genomics offers a comprehensive service integrating CRISPR screening with high-throughput phenotypic analysis. This service is versatile, addressing various research areas and issues effectively. It encompasses the entire process from sgRNA library construction to the screening and validation of functional genes or drug targets.
Key Considerations in CRISPR/Cas9 Screen Design
Designing CRISPR/Cas9 screens involves addressing several critical considerations that directly impact the accuracy and efficacy of subsequent experimental outcomes. CRISPR library screening and library design represent pivotal stages in the CRISPR application domain, influencing the quality and reliability of experimental results.
Selection of CRISPR sgRNA Libraries
Choosing between pooled sgRNA libraries and arrayed sgRNA libraries is fundamental. This decision dictates the types of questions that can be addressed by CRISPR library screens and influences the reliability of resultant data. Additionally, the choice between whole-genome libraries and targeted libraries aimed at specific genes or pathways must be deliberated.
Selection of Cell Lines for Screening
Careful consideration should be given to the selection of appropriate cell lines for CRISPR/Cas9 screening. The chosen cell line should be well-suited to the experimental objectives, ensuring robustness in detecting sgRNA-mediated gene editing effects.
Data Analysis and Hit Validation
Effective strategies for data analysis and hit validation are critical components of CRISPR/Cas9 screen design. Rigorous data analysis methodologies are essential for identifying genuine hits, while subsequent validation steps are necessary to ascertain the biological relevance of identified hits.
The decisions made in each of these areas collectively define the scope of questions that can be addressed by CRISPR library screens and determine the credibility of the resulting data. This article by CD Genomics aims to provide an informative overview of CRISPR library customization and screening, offering insights to enhance understanding and application of this transformative technology.
Designing sgRNA Libraries
Efficient planning of CRISPR screening begins with the design of sgRNA libraries targeting genes or loci of interest. Subsequently, oligonucleotide pools corresponding to the sgRNA designs are synthesized and cloned into vectors for sgRNA expression, or transcribed into RNA for transfection into the cellular systems undergoing screening.
The rational and efficient design of sgRNA libraries is pivotal for genome/metabolic pathway editing. Combined with high-throughput screening technologies, sgRNA libraries play a crucial role in drug development, gene function studies, molecular diagnostics, disease therapy, and crop breeding.
This approach underscores the foundational importance of well-designed sgRNA libraries in advancing applications across diverse scientific domains.
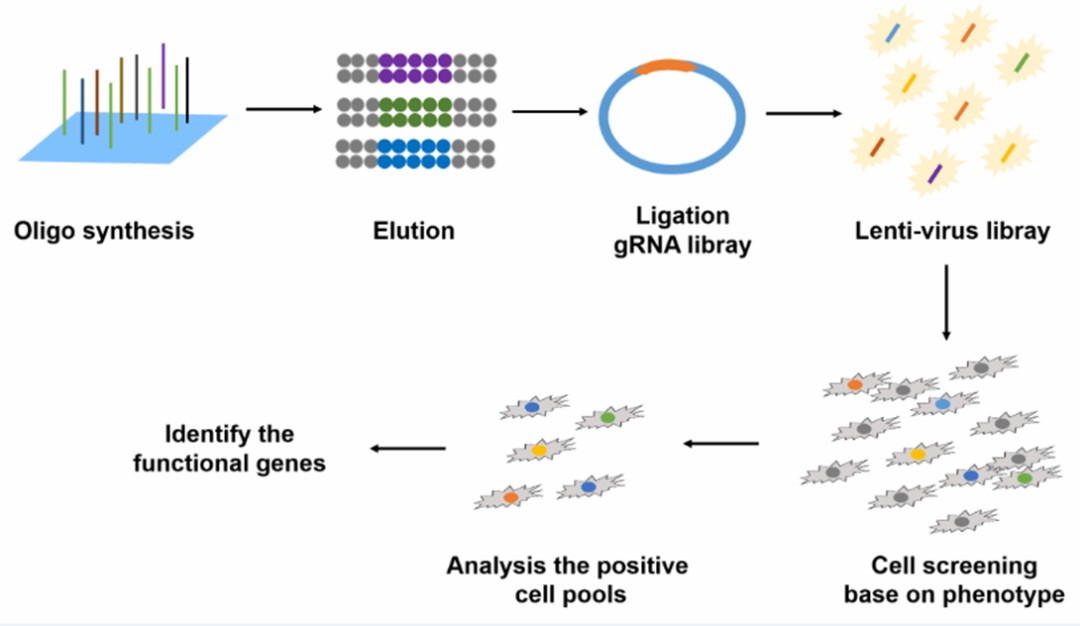 Figure 2. CRISPR library construction and downstream work
Figure 2. CRISPR library construction and downstream work
Types of sgRNA Libraries
sgRNA libraries encompass various types, including but not limited to whole-genome libraries, lncRNA libraries, and specific libraries targeting signal pathways, cell apoptosis, cell proliferation, ion channels, nuclear receptors, and various disease-related libraries.
Construction of Whole-Genome sgRNA Libraries
Whole-genome sgRNA libraries have broad applications, allowing for the design and screening of diverse types of genomic DNA. This includes ORF cDNA, lncRNA cDNA, and specific regions of cDNA fragments. Efficient construction of full-length cDNA and genome DNA sgRNA libraries provides robust tools and methodologies for high-throughput functional gene screening and research into relevant drug targets.
Software Tools for CRISPR sgRNA Design
Following the selection of target genes and Cas nucleases, the design of specific guide RNA sequences is crucial. To ensure minimal off-target effects and maximum on-target efficiency, a range of software tools are available to aid in optimal guide RNA design. The following are among the most popular guide RNA design tools currently available on the market:
| Synthego Design Too |
Cas-OFFinder |
| Broad Institute GPP sgRNA Designer |
CRISPR-Era |
| CRISPOR |
Benchling CRISPR Guide RNA Design tool |
| CHOPCHOP |
E-CRISP |
| Off-Spotter |
|
Key Considerations and Limitations in sgRNA Design
When designing sgRNA for CRISPR experiments, several critical factors must be carefully considered:
Firstly, the GC content of sgRNA is crucial for its stability, typically ranging from 40% to 80%. Maintaining this range ensures structural stability and functional reliability.
Secondly, the length of sgRNA must strictly fall between 17 to 24 nucleotides, adjusted according to the specific type of Cas nuclease used. Shorter sequences aid in reducing off-target effects, but excessively short sequences may compromise efficiency, necessitating optimal length balance.
Additionally, mismatches between gRNA and the target site significantly influence the extent of off-target effects, depending on their number and specific positions. Therefore, minimizing mismatches with the target site during sgRNA design is essential to mitigate the risk of off-target effects.
Finally, due to the unpredictable activity and specificity of sgRNA, it is advisable to design multiple sgRNAs for each gene of interest. This approach facilitates the selection of the most effective candidates during experimental screening, ensuring the reliability and accuracy of the experiments and enhancing the success rate of CRISPR editing technologies.
This refined text emphasizes the meticulous considerations and strategic planning required in sgRNA design for CRISPR experiments, reflecting a scientific academic style with an emphasis on precision and reliability in genetic editing techniques.
High-Throughput sgRNA Library Screening
High-throughput sgRNA library screening is a scientific method aimed at functional screening and enrichment within specific cells using whole-genome or pathway-specific sgRNA libraries. This approach involves subsequent PCR amplification and deep sequencing analysis to uncover genes associated with specific phenotypes or explore potential novel drug targets at the genome-wide level. The core process of CRISPR/Cas9 gRNA library screening comprises five key steps: initial selection of appropriate gRNA libraries, followed by library amplification, lentiviral packaging of the amplified gRNA library, screening of the gRNA library in target cell lines, and analysis through PCR amplification and NGS sequencing technologies. Common screening techniques include CRISPR-based knockout, activation, and inhibition screens, collectively forming the comprehensive framework of high-throughput sgRNA library screening.
This refined text articulates the structured approach and scientific rigor involved in high-throughput sgRNA library screening, focusing on its methodological framework and application in genetic and drug target discovery.
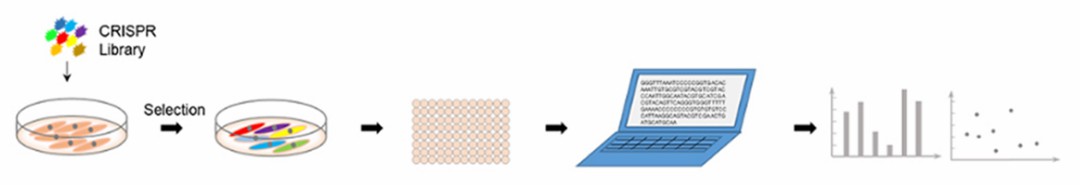 Figure 3. High-throughput sgRNA library screening process
Figure 3. High-throughput sgRNA library screening process
CRISPR Knockout Screening
CRISPR-Cas9 technology enables precise genome manipulation through the induction of irreversible gene disruption via non-homologous end joining (NHEJ) or homology-directed repair (HDR) mechanisms. Subsequent screening identifies phenotype variations induced by these disruptions, facilitating detailed analysis and research.
| Application |
Diminished vitality, increased drug sensitivity, reduced proliferation, decreased migration capability |
| Advantages |
|
|
- Low noise |
|
- Facilitates detection of essential survival genes |
|
- Higher sensitivity compared to previous RNAi platforms |
|
- Ability to target nearly the entire genome, including non-coding regions |
| Disadvantages |
|
|
- Low cleavage efficiency |
|
- Occurrence of off-target effects |
|
- Requires increased number of sgRNAs to ensure effectiveness for each target |
|
- Heterogeneity and heterozygous knockouts observed |
|
- Potential induction of cytotoxicity due to increased double-strand breaks (DSBs) in the genome |
CRISPR Activation Screening
Utilizing CRISPR-dCas9 technology enables reversible genome-wide gene activation without disrupting genomic sequences. This process typically involves introducing additional regulatory domains followed by meticulous phenotype screening.
| Application |
Modulation of promoter regions to activate or overexpress genes or non-coding elements. Functional gain analysis provides insights into drug-resistant genes or essential proteins. |
| Advantages |
|
|
- No disruption of genomic sequences |
|
- Superior to previous cDNA library overexpression methods |
|
- Enhanced feasibility of lncRNA expression through promoter modulation |
|
- Robust in vivo activation models |
| Disadvantages |
|
|
- Susceptible to sequence variability |
|
- Requires larger Cas9 complexes |
|
- Challenges with AAV packaging due to its size; lentiviruses and adenoviruses may provoke host responses. |
CRISPR Suppression Screening
CRISPR-dCas9 technology facilitates reversible genome-wide gene suppression without altering genomic sequences. This process typically involves introducing additional regulatory domains followed by screening for resultant phenotypic changes.
| Application |
Detection of functional deficits in populations. Enables precise targeting through various functional suppression complexes. |
| Advantages |
|
|
- No disruption of genomic sequences |
|
- Absence of off-target cytotoxicity |
|
- Precision in interfering with regulatory elements across the genome |
|
- Effective knockdown of lncRNAs |
| Disadvantages |
|
|
- Susceptibility to sequence variability |
|
- Suboptimal regulation of complex transcription start sites (TSS) |
|
- Requirement for larger Cas9 complexes |
|
- Challenges in packaging due to large size; lentiviruses and adenoviruses may provoke host responses. |
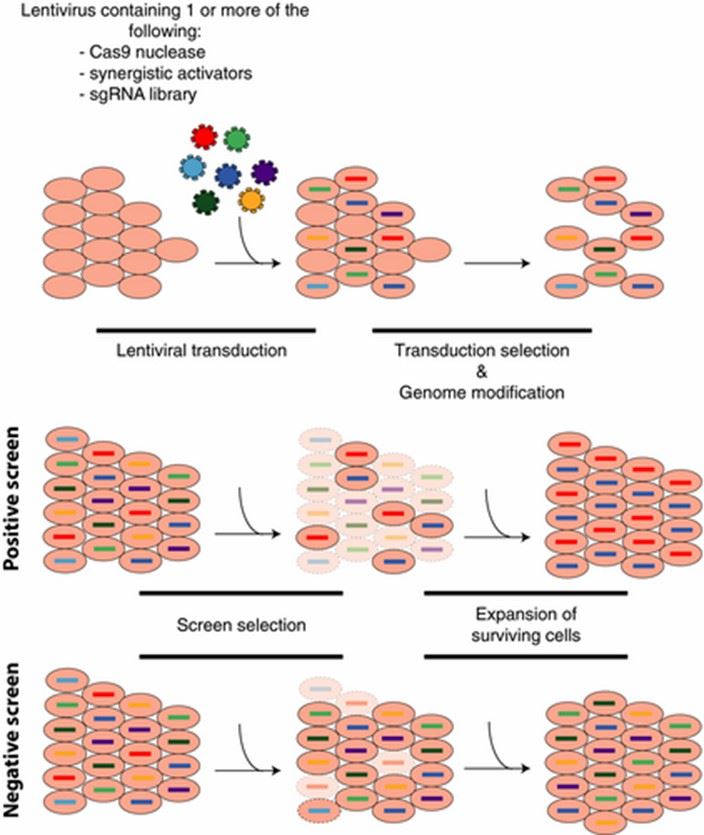 Figure 4. Types of CRISPR screens
Figure 4. Types of CRISPR screens
CRISPR Screening Design
Drug pressure screening is a method employing high-throughput sequencing to systematically compare the types and abundances of sgRNAs between control and experimental groups. This approach accurately evaluates the impact of gene knockout on cellular drug tolerance or sensitivity.
Cell sorting methods rely on FACS technology to precisely isolate target cell populations. They facilitate in-depth comparison of sgRNA abundances among different populations, thereby enabling efficient selection of functional sgRNAs.
Single-cell CRISPR screening integrates CRISPR screening technology with single-cell transcriptome sequencing (scRNA-seq). This synergy reveals the functional characteristics of genes and genetic regulatory networks, providing robust support for in-depth research.
Cell Line Selection for Genome-Wide Screening
Following the selection of a specific CRISPR/Cas9 library, the subsequent critical decision revolves around choosing appropriate cell lines. Numerous factors influence the selection of cell lines for genome-wide screening. To mitigate specific issues such as genetic background disparities or varied transduction efficiencies, it is advisable to conduct parallel screening across multiple cell lines.
When selecting cell lines for CRISPR library screening, adherence to the following principles is recommended: Firstly, consideration should be given to the gene copy number variation among cell lines. Diploid cell lines may yield screening results with higher signal-to-noise ratios and greater reliability compared to hyperdiploid counterparts. Secondly, attention should be focused on the activity status of DNA repair pathways within specific cell lines, particularly any deficiencies in the HDR pathway. Additionally, the sensitivity of cell lines to the selection agents used during screening is crucial and should not be overlooked. The choice of cell lines directly impacts the number and types of genes identifiable during the screening process. Lastly, the transduction efficiency of cell lines is a critical determinant of screening success and should be meticulously evaluated.
Analysis of Screening Results
During CRISPR library screening, to ensure accuracy and reliability of data, deep sequencing analysis of samples is typically conducted using PCR amplification techniques. The design of PCR primers is crucial in this process, ensuring specific amplification of the lentiviral backbone containing sgRNAs to precisely capture target information. Moreover, to maintain complexity of sgRNAs and introduce significant library diversity, a staggered sequencing primer design is employed to achieve comprehensive coverage of the library.
At different stages of screening, cell pellets from various time points are collected to extract DNA for subsequent analysis. To ensure sample quality and data reliability, extraction procedures must carefully control proportions to avoid overloading and minimize the risk of sample diversity loss.
To ensure representativeness and reliability of screening results, the cell populations screened and analyzed should have sufficient numbers. Typically, inadequately represented sgRNAs require capture from sufficiently large cell populations, with representativeness generally targeted at 300-1000-fold coverage. Furthermore, the screening process typically involves operations across multiple culture plates/flasks and several hours of tissue culture on seeding and harvest days to ensure smooth experiment execution and result accuracy.
Subsequent Steps in CRISPR Library Screening
The downstream processes of CRISPR library screening encompass several pivotal stages. Initially, each identified candidate target is validated to ensure accuracy and reliability. Subsequently, orthogonal screening methods are employed to distinguish true positives from false positives, thereby enhancing screening precision. Following this, supplementary screening is conducted to further validate hit rates and identify the most promising candidate targets. Subsequent steps involve the establishment of cell lines corresponding to potential protein knockouts, laying the groundwork for subsequent investigations. Additionally, cDNA overexpression techniques are utilized to simulate gene transcription activation processes, facilitating exploration of gene functions and regulatory mechanisms. Finally, validation includes modification verification at the individual genetic and/or transcriptional levels to confirm the reliability and efficacy of screening results. These validation procedures vary depending on the type of screening, aiming to ensure the accuracy and reliability of screening outcomes.
Reference:
- Miles, L. A., Garippa, R. J., & Poirier, J. T. (2016). Design, execution, and analysis of pooledin vitroCRISPR/Cas9 screens. The FEBS Journal, 283(17), 3170-3180.


 Sample Submission Guidelines
Sample Submission Guidelines
 Figure 1. CRISPR gene editing technology
Figure 1. CRISPR gene editing technology Figure 2. CRISPR library construction and downstream work
Figure 2. CRISPR library construction and downstream work Figure 3. High-throughput sgRNA library screening process
Figure 3. High-throughput sgRNA library screening process Figure 4. Types of CRISPR screens
Figure 4. Types of CRISPR screens