Circulating Tumor DNA (ctDNA) Sequencing in Cancer Research
Currently, despite the approval of multiple targeted therapies for the treatment of solid tumors, the challenge of acquired resistance persists, significantly diminishing their efficacy. While high-throughput analysis of tumor tissue samples can reveal polyclonal resistance patterns, biopsies taken after resistance emerges are less practical and fail to capture the genetic diversity within patients. In this paper, the authors delve into the characterization of secondary drug resistance mechanisms in cancer patients. They employ next-generation sequencing technology to analyze circulating tumor DNA (ctDNA), showcasing the immense potential of ctDNA sequencing in guiding therapeutic approaches and enhancing prognostic accuracy in clinical practice.
The advent of targeted therapies has revolutionized tumor patient care. Notably, early efforts targeted hormone receptors, while more recent therapies focus on mutated or overexpressed kinases. These advancements have positively impacted patient prognosis across various tumor types, such as non-small cell lung cancer, melanoma, and colorectal cancer. However, the prevalent issue of acquired resistance, often emerging after months or even years of treatment, poses a formidable challenge. Key mechanisms behind this resistance involve secondary alterations in the drug target and mutations in alternative components of the oncogene-induced signaling pathway. Despite their significance, the true incidence of these mechanisms and their impact on patient outcomes remains elusive.
Circulating tumor DNA (ctDNA) sequencing emerges as a pivotal tool for cancer genomic profiling due to its widespread application, reproducibility, and feasibility. By providing actionable mutational targets for patients grappling with drug resistance, ctDNA sequencing stands as a promising avenue for advancing precision medicine in oncology.
Alterations in Overall Characterization and Drug Resistance
In the examination of the STING cohort, researchers employed circulating tumor DNA (ctDNA) molecular profiling to scrutinize the prevalence of genomic aberrations linked to acquired drug resistance and their impact on the prognostic outlook for patients undergoing targeted therapy in advanced stages. By comparing circulating free DNA (cfDNA) sequencing data with pre-treatment tumor tissue sequencing data, the study aimed to pinpoint potential mechanisms responsible for acquired resistance.
A total of 626 patients with advanced cancer, enrolled in the Institutional Molecular Profiling Program (STING, NCT04932525) and subjected to targeted therapy, were comprehensively assessed to derive ctDNA profile-based molecular profiles. Among these patients, 193 (31%) were identified to harbor at least one previously validated resistance alteration, with 86 patients (14%) exhibiting more than one detectable resistance alteration. This finding underscores the inherent tumor heterogeneity associated with acquired resistance.
Furthermore, patients manifesting at least one detectable ctDNA alteration experienced a significantly reduced overall survival (OS) compared to those devoid of any resistance mechanism—16.2 months (95% CI 14.8-18.3) versus 10.2 months (6.5-13.9) (p < 0.001). These results underscore the clinical relevance of ctDNA profiling in uncovering resistance mechanisms and its potential implications for patient outcomes in the context of targeted therapy for advanced-stage cancers.
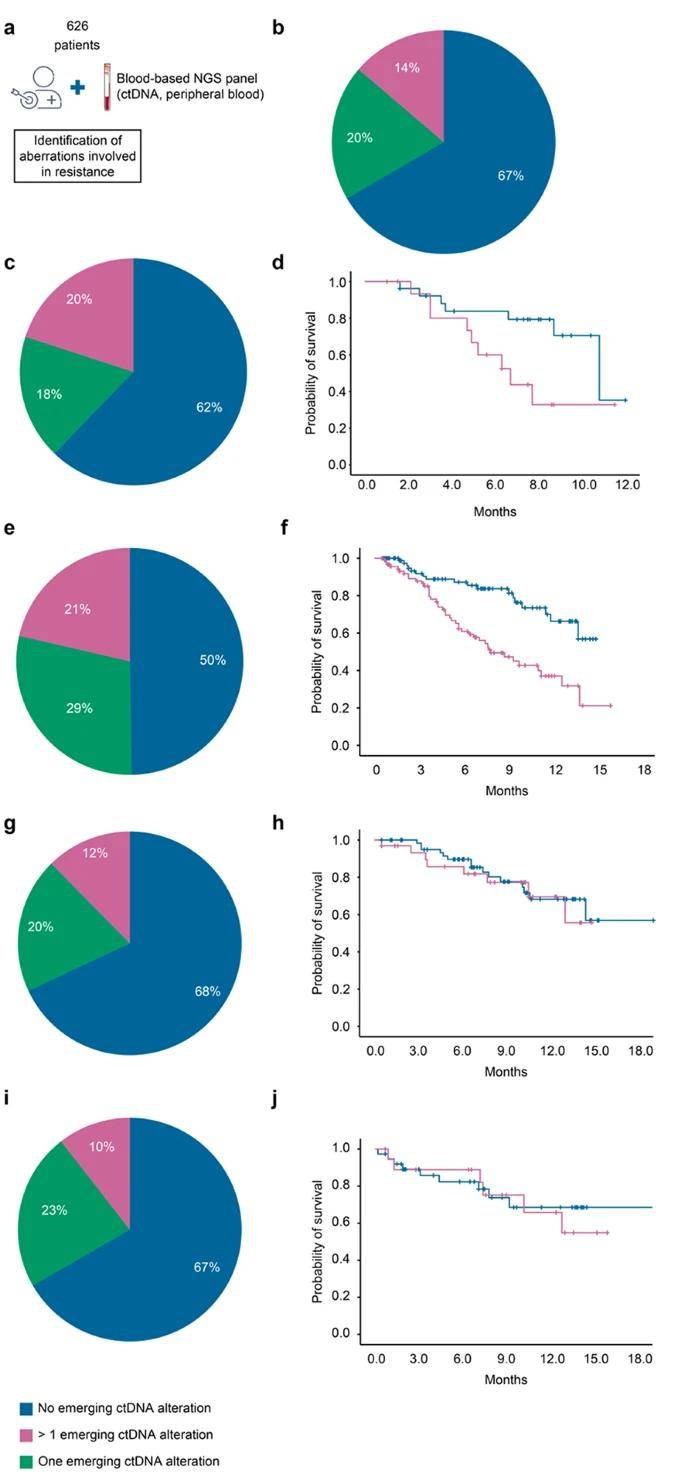 Incidence of ctDNA emerging alterations in patients with advanced cancer and treated with targeted therapy. (Bayle et al., 2023)
Incidence of ctDNA emerging alterations in patients with advanced cancer and treated with targeted therapy. (Bayle et al., 2023)
Reference:
- Bayle, A., Belcaid, L., Palmieri, LJ. et al. Circulating tumor DNA landscape and prognostic impact of acquired resistance to targeted therapies in cancer patients: a national center for precision medicine (PRISM) study. Mol Cancer 22, 176 (2023).
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.


 Sample Submission Guidelines
Sample Submission Guidelines
 Incidence of ctDNA emerging alterations in patients with advanced cancer and treated with targeted therapy. (Bayle et al., 2023)
Incidence of ctDNA emerging alterations in patients with advanced cancer and treated with targeted therapy. (Bayle et al., 2023)