In microbiome research (including metagenomics and 16S rRNA sequencing), the terms absolute abundance and relative abundance are frequently encountered. However, what exactly do these terms mean, and why is it important to differentiate between them?
What is Relative Abundance?
Relative abundance refers to the proportion of a specific microorganism within the entire microbial community. In other words, it does not provide the actual number of microorganisms but rather indicates the proportion of that microorganism relative to the total microbial count. The sum of relative abundances typically equals 100% (or 1).
Example:
Assume that in a sample, a total of 300,000 bacteria are detected, with 100,000 being species A and 200,000 being species B. The relative abundance can be calculated as follows:
- Relative abundance of species A = 100,000 / 300,000 = 33.33%
- Relative abundance of species B = 200,000 / 300,000 = 66.67%
Relative abundance is relatively straightforward to calculate, and since it is normalized to the total microbial count, it is unaffected by the total sample size. High-throughput sequencing techniques, such as 16S rRNA sequencing, are commonly used to obtain relative abundance data.
What is Absolute Abundance?
Absolute abundance refers to the actual number of a specific microorganism present in a sample. It is typically quantified as the "number of microbial cells per gram/milliliter of sample." This measure directly informs us about the actual quantity of microorganisms in the sample.
Example:
In a water sample, suppose 100,000 bacteria of species A and 200,000 bacteria of species B are detected. The absolute abundance of species A is 100,000 cells, and for species B, it is 200,000 cells.
Absolute abundance data is usually obtained through quantitative techniques such as quantitative PCR (qPCR), which require additional experimental steps and precise measurement tools.
Key Differences Between Absolute and Relative Abundance
The main differences between absolute and relative abundance are as follows:
- Absolute abundance provides the actual count of microorganisms, which reflects the true number of microbes in the sample.
- Relative abundance describes the proportional relationship between different microorganisms within a sample, allowing for comparison of their relative distributions.
However, a limitation of relative abundance is that it may not accurately reflect the true changes in a microorganism's abundance when the total sample size varies. For instance, if the numbers of both species A and species B decrease proportionally, the relative abundance might remain unchanged, even though the actual number of these microorganisms has decreased. In contrast, absolute abundance would reveal the actual decrease in microbial numbers.
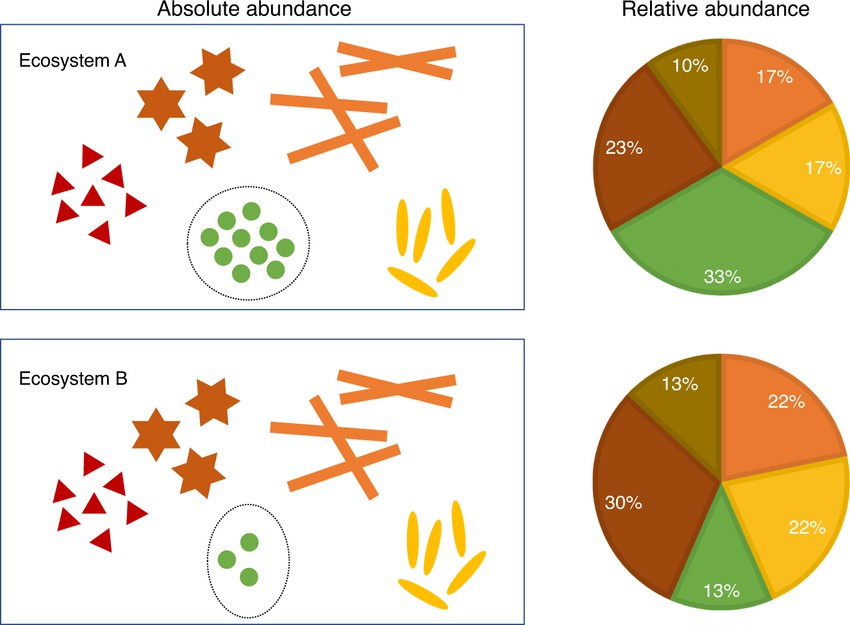 Figure 1. The distinction between absolute abundances and relative abundances (Huang Lin et al., 2020)
Figure 1. The distinction between absolute abundances and relative abundances (Huang Lin et al., 2020)
When to Use Absolute Abundance and When to Use Relative Abundance?
Absolute abundance: If the goal is to determine the actual number of microorganisms (such as in disease monitoring or precise quantification of microbial load), absolute abundance is more reliable.
Relative abundance: If the focus is on understanding the community structure and comparing the proportions of different microorganisms within a sample (such as in ecological studies of microbial populations), relative abundance is often preferred. This approach highlights the proportional relationships among microbes within the community.
By understanding these two different approaches, researchers can select the appropriate method for their specific study objectives, ensuring that the data obtained provides meaningful and accurate insights into the microbiome.
Absolute and Relative Abundance in 16S rRNA Sequencing
16S rRNA sequencing is a widely employed technique for analyzing microbial community structure. It works by amplifying the 16S rRNA gene of bacteria and archaea, which helps identify the types of microorganisms present in a sample.
In 16S sequencing, relative abundance is commonly used. This is because the sequencing results typically provide the sequence reads for each bacterial taxon (e.g., operational taxonomic units (OTUs) or amplicon sequence variants (ASVs)), rather than the actual quantity of organisms. Variations in sequencing depth and efficiency can influence the total number of sequences across different samples, prompting the conversion of sequence counts into relative abundance for comparison between samples.
To determine absolute abundance in 16S rRNA sequencing, additional methods, such as qPCR or flow cytometry, are required to quantify the total microbial load in the sample. The absolute abundance of each microbial species can then be calculated by multiplying the relative abundance by the total microbial quantity.
Example:
If qPCR reveals that the total bacterial count in a sample is 1 million, and the relative abundance of species A is 20%, the absolute abundance of species A would be:
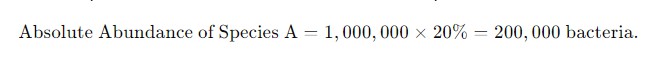 Figure 2. The formula for calculating absolute abundance
Figure 2. The formula for calculating absolute abundance
Summary
In 16S sequencing, relative abundance is the primary method of analysis because it eliminates the variability caused by differences in sequencing depth. If absolute abundance is required, quantitative techniques must be incorporated to supplement the data.
Absolute and Relative Abundance in Metagenomic Sequencing
Metagenomic sequencing involves directly sequencing the genomes of all microorganisms present in a sample, providing a more comprehensive analysis. This method allows for the detection of bacteria, fungi, viruses, and other microbial genetic information. Metagenomic sequencing offers higher resolution and enables direct insights into microbial functional characteristics.
Similar to 16S rRNA sequencing, metagenomic sequencing typically utilizes relative abundance for data analysis. This is due to the fact that the total number of sequence reads in metagenomic sequencing can be influenced by sequencing depth, leading to significant variation in the total read count between samples. Therefore, the use of relative abundance ensures comparability across samples.
Absolute Abundance in Metagenomics
To determine the absolute abundance of each microbial species in metagenomic sequencing, total microbial abundance data for the sample is required. As with 16S rRNA sequencing, methods such as qPCR or other quantitative techniques can be employed to estimate total microbial load. The absolute abundance for each microorganism can then be calculated by multiplying the relative abundance by the estimated total abundance.
Metagenomics vs. 16S rRNA Sequencing: Abundance Metrics
16S rRNA Sequencing: This method is more suitable for quickly assessing the composition and changes in microbial communities, especially when budget constraints exist. The calculation of relative abundance is straightforward; however, the resolution is limited due to the sequencing of only a specific fragment of the 16S gene, making it challenging to obtain accurate absolute counts.
Metagenomic Sequencing: This approach captures a broader range of microbial taxa, including bacteria, viruses, fungi, and other organisms, while also providing more in-depth analysis of microbial functional traits. Although metagenomic sequencing is more costly, it offers richer information, including gene functions and ecological roles. Like 16S rRNA sequencing, metagenomic sequencing primarily relies on relative abundance analysis unless supplemented with quantitative techniques to derive absolute abundance.
Conversion Between Absolute and Relative Abundance
Converting Absolute Abundance to Relative Abundance
It is straightforward to convert absolute abundance to relative abundance by dividing the absolute abundance of each species by the total absolute abundance of all species in the sample.
Formula:
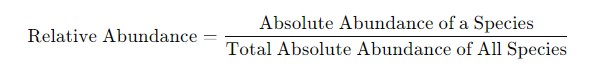 Figure 3. The formula for converting Absolute Abundance to Relative Abundance
Figure 3. The formula for converting Absolute Abundance to Relative Abundance
To convert relative abundance to absolute abundance, the total microbial abundance in the sample must be known. The absolute abundance can then be calculated by multiplying the relative abundance by the total microbial abundance.
Formula:
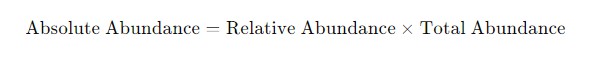 Figure 4. The formula for converting relative abundance to absolute abundance
Figure 4. The formula for converting relative abundance to absolute abundance
By understanding these methods of abundance calculation and conversion, researchers can select the appropriate approach for their specific experimental design, ensuring the accuracy and comparability of microbial community data across various study types.
Code Example for Converting Abundance Data in 16S rRNA or Metagenomic Sequencing Using R
In both 16S rRNA and metagenomic sequencing, the fundamental approach for calculating relative and absolute abundance remains consistent. Below is an example demonstrating how to perform these conversions in R:
1. Converting Absolute Abundance to Relative Abundance in 16S rRNA or Metagenomic Sequencing
Suppose we have absolute abundance data for a sample, where each row represents a sample and each column corresponds to a different microorganism:
# Absolute abundance data (rows = samples, columns = microorganisms)
absolute_abundance <- matrix(c(500, 1000, 200,
800, 400, 600,
300, 500, 900),
nrow = 3, byrow = TRUE)
# Calculate the total abundance for each sample
total_abundance <- rowSums(absolute_abundance)
# Calculate relative abundance
relative_abundance <- absolute_abundance / total_abundance
# Output relative abundance matrix
relative_abundance
2. Converting Relative Abundance to Absolute Abundance (Requires Total Sample Abundance)
Suppose we have a relative abundance matrix, and the total microbial abundance for each sample is known (obtained via methods such as qPCR):
# Relative abundance matrix (rows = samples, columns = microorganisms)
relative_abundance <- matrix(c(0.2, 0.4, 0.1,
0.4, 0.2, 0.3,
0.3, 0.25, 0.45),
nrow = 3, byrow = TRUE)
# Total microbial abundance per sample (obtained via qPCR or similar methods)
total_abundance <- c(1000, 1500, 2000)
# Calculate absolute abundance
absolute_abundance <- relative_abundance * total_abundance
# Output absolute abundance matrix
absolute_abundance
Are the Reads from Alignment Considered Absolute Abundance?
The number of reads obtained from sequence alignment cannot be directly equated to absolute abundance. Rather, these reads represent the sequence counts for a particular microorganism in a sample. This value does not correspond to the actual microbial quantity (absolute abundance) due to several influencing factors, including:
- Sequencing Depth: Different samples may have varying sequencing depths, which affect the number of reads generated. Samples with higher sequencing depth will produce more reads.
- PCR Amplification Bias: In amplification-based sequencing techniques (e.g., 16S rRNA sequencing), certain microbial gene fragments may be preferentially amplified, leading to an overrepresentation of certain taxa and potential errors in the relative abundance estimation.
- Genome Size: In metagenomic sequencing, the genome size of different microorganisms can vary significantly. Microorganisms with larger genomes are likely to produce more reads than those with smaller genomes.
Thus, the number of reads is generally considered an approximation of relative abundance, reflecting the "relative presence" of a microorganism in the microbial community after sequencing.
To convert read counts to absolute abundance, additional quantitative steps are required. For instance, methods such as qPCR or flow cytometry can be employed to determine the total microbial abundance in the sample. Once the total abundance is known, absolute abundance for each microorganism can be calculated by combining this data with the read counts.
Conversion Formula:
The following formula allows the conversion of relative sequencing data to actual microbial quantities, thus enabling the calculation of absolute abundance:
 Figure 5. The formula for converting reads count to absolute abundance
Figure 5. The formula for converting reads count to absolute abundance
The Importance of Absolute Quantification in Microbiome Research
An increasing body of evidence suggests that measuring the absolute abundance of microorganisms can provide more comprehensive data compared to relative quantification, offering the potential to correct certain erroneous conclusions derived from relative abundance data. Moreover, experimental biases introduced during sample processing can significantly affect the accurate measurement of microbiome composition. Systematic evaluations of sample preservation methods and the impact of different storage temperatures on microbial communities are necessary to better understand the sources of analytical biases. However, existing literature presents inconsistent conclusions on this matter, highlighting the need for further investigation to clarify these discrepancies.
Incorporating absolute quantification into the assessment of microbiome samples can enhance the precision of microbial abundance measurements. This approach is critical for identifying and addressing systematic biases arising from variations in experimental conditions, such as differences in sample preservation solutions and storage temperatures. The recent study published in Nature Biotechnology, titled Quantifying Bias Introduced by Sample Collection in Relative and Absolute Microbiome Measurements , provides a comprehensive evaluation of the biases introduced during sample collection, comparing both relative and absolute quantification methods. In this study, the authors assessed how two commonly used preservation solutions and short-term storage at different temperatures influence both relative and absolute microbial abundances.
The authors strongly advocate for the incorporation of absolute quantification in microbiome research. They emphasize that the lack of a universally accepted absolute quantification method in microbiome studies is a notable shortcoming. Absolute quantification is critical as it prevents the erroneous correlations between microorganisms and host biology that may arise when only relative abundance data are considered. Furthermore, it significantly alters conclusions regarding the interactions between microbiome components and their influence on the human host. Thus, the adoption of absolute abundance measurements is essential for more accurate interpretations of microbiome data.
Absolute Abundance Quantification Workflow
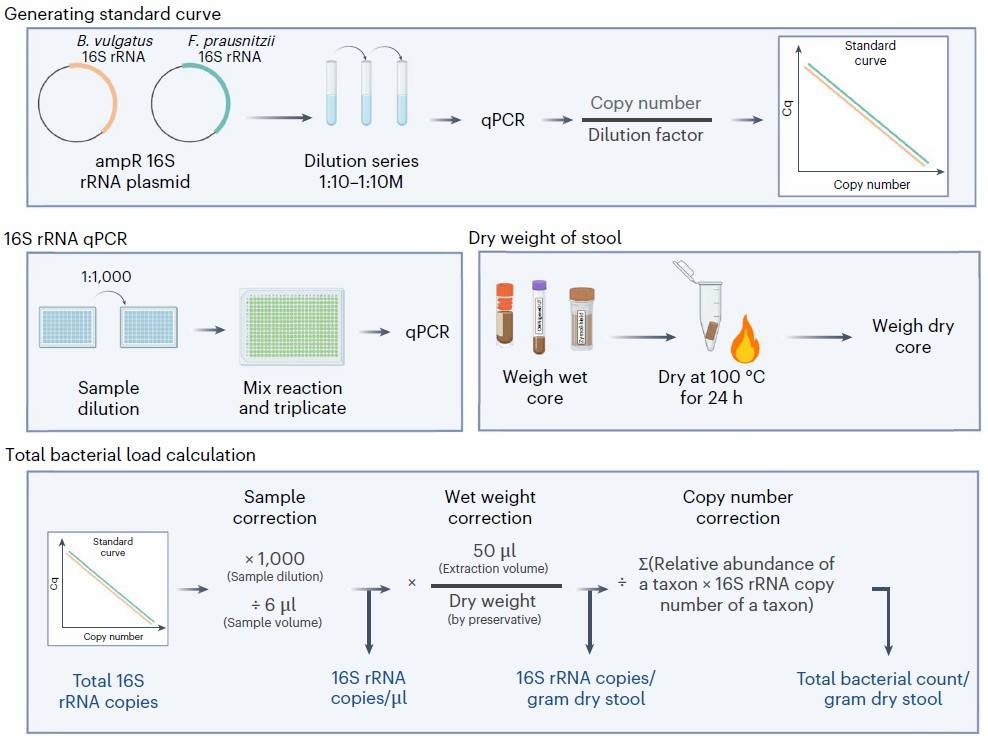 Figure 6. Workflow of absolute abundance quantification in Microbiome
Figure 6. Workflow of absolute abundance quantification in Microbiome
Integration of Relative and Absolute Abundance Data in Microbiome
Estimating the heritability of microbial taxa remains a complex challenge in microbiome research. Traditionally, relative abundance data have been employed to infer heritability; however, the limitations intrinsic to relative abundance measurements can result in inaccurate or misleading estimates. These limitations stem from the fact that relative abundance does not necessarily reflect the true genetic contributions of microbial communities, as it is influenced by factors such as sequencing depth and sample variability.
Relative abundance data can misrepresentheritability of the microbiome presents an innovative approach for estimating the heritability of microbiomes by integrating both relative and absolute abundance data. The proposed method entails the concurrent analysis of relative abundance alongside absolute abundance to provide a more precise estimate of heritability. Initially, relative abundance data are utilized to estimate the heritability of microbial taxa. Following this, the inclusion of absolute abundance data enables a more accurate assessment, mitigating the biases inherent in relative abundance alone. This integrated approach offers a more reliable representation of microbiome heritability, yielding insights that are closer to the actual genetic influence of microbial communities.
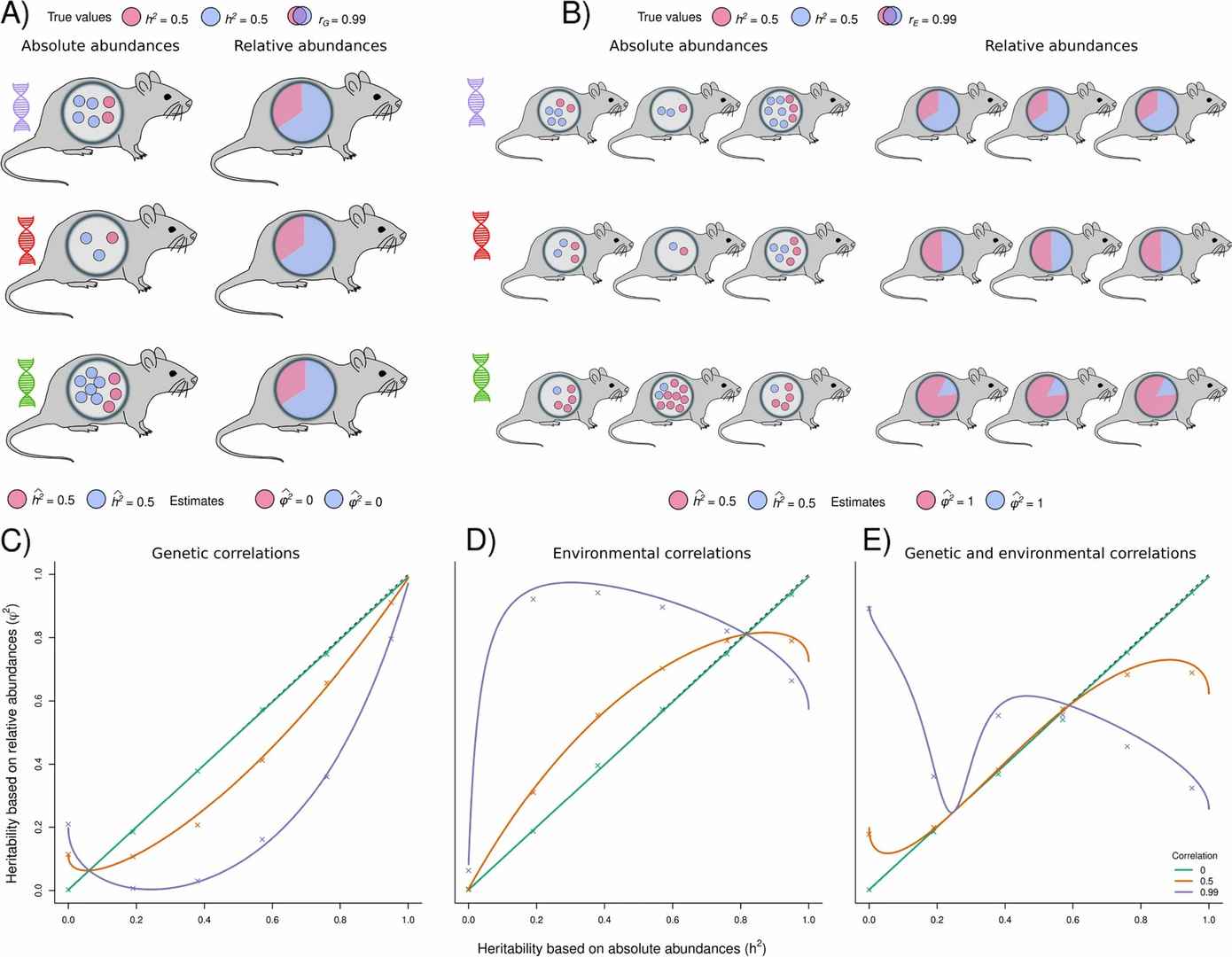 Figure 7. The use of relative abundances leads to biased heritability estimates when there exists host genetic and/or environmental correlations between microbes.
Figure 7. The use of relative abundances leads to biased heritability estimates when there exists host genetic and/or environmental correlations between microbes.
Comprehensive Data Acquisition: The technology enables the simultaneous generation of three distinct datasets from a single test, thereby providing a richer and more detailed result.
Minimized Sample Requirements: The method requires a lower sample volume, reducing the risk of data loss due to insufficient sample availability or the absence of backups.
High Throughput and Sensitivity: The technology offers high throughput, with the ability to achieve absolute quantification for a wide range of microbial species within the detection range, ensuring comprehensive analysis.
Complementary Relative and Absolute Quantification: The combination of relative and absolute quantification provides a robust validation of results, reducing the occurrence of false positives commonly associated with traditional relative quantification methods.
Elimination of Cross-Platform qPCR Systematic Errors: The approach mitigates systematic biases often encountered in cross-platform qPCR quantification, ensuring more reliable results.
Superior Specificity and Sensitivity: The internal standard method offers higher specificity, sensitivity, and reproducibility in quantification compared to conventional qPCR techniques.
Minimized PCR Inhibition: The method reduces the impact of residual PCR inhibitors from DNA extraction processes, ensuring the accuracy and reliability of the results.
Simplified Primer Design and Optimization: The approach avoids the challenges typically encountered in primer design and optimization that are inherent to qPCR and other quantitative assays.
Summary
Absolute and relative abundance are two essential concepts in microbiome research. Absolute abundance provides the actual count of microorganisms in a sample, while relative abundance describes the proportional representation of each microorganism within the sample.
In the context of 16S rRNA and metagenomic sequencing, both types of abundance have specific applications:
- 16S rRNA Sequencing: Relative abundance is primarily used for community structure analysis. If absolute abundance is needed, it must be determined through supplementary techniques, such as qPCR, to quantify the total microbial load.
- Metagenomic Sequencing: While relative abundance is used to gain a comprehensive understanding of microbial communities and their functions, absolute abundance requires quantitative methods to estimate total microbial abundance.
This article aims to clarify the abundance-related concepts in these sequencing technologies and provide guidance on how to apply both absolute and relative abundance effectively in microbiome studies.
References:
-
Maghini, D.G., Dvorak, M., Dahlen, A. et al. Quantifying bias introduced by sample collection in relative and absolute microbiome measurements. Nat Biotechnol 42, 328–338 (2024). https://doi.org/10.1038/s41587-023-01754-3
- Bruijning, M., Ayroles, J.F., Henry, L.P. et al. Relative abundance data can misrepresent heritability of the microbiome. Microbiome 11, 222 (2023). https://doi.org/10.1186/s40168-023-01669-w


 Sample Submission Guidelines
Sample Submission Guidelines
 Figure 1. The distinction between absolute abundances and relative abundances (Huang Lin et al., 2020)
Figure 1. The distinction between absolute abundances and relative abundances (Huang Lin et al., 2020)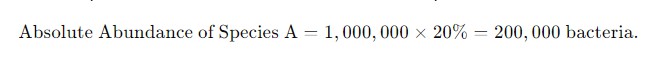 Figure 2. The formula for calculating absolute abundance
Figure 2. The formula for calculating absolute abundance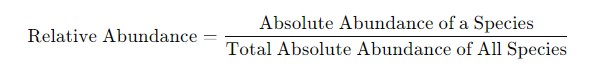 Figure 3. The formula for converting Absolute Abundance to Relative Abundance
Figure 3. The formula for converting Absolute Abundance to Relative Abundance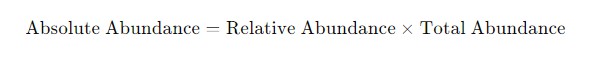 Figure 4. The formula for converting relative abundance to absolute abundance
Figure 4. The formula for converting relative abundance to absolute abundance Figure 5. The formula for converting reads count to absolute abundance
Figure 5. The formula for converting reads count to absolute abundance Figure 6. Workflow of absolute abundance quantification in Microbiome
Figure 6. Workflow of absolute abundance quantification in Microbiome Figure 7. The use of relative abundances leads to biased heritability estimates when there exists host genetic and/or environmental correlations between microbes.
Figure 7. The use of relative abundances leads to biased heritability estimates when there exists host genetic and/or environmental correlations between microbes.