Metagenomics is a new generation of high-throughput sequencing (NGS) technology that takes the microbial population genome in a specific environment as the research object. Based on the analysis of microbial diversity, population structure and evolutionary relationship, it can further explore the functional activity of microbial population, the relationship between mutual collaboration and the environment, and explore the potential biological significance.
Metagenomics for studying the pathogenesis of infectious diseases
Infection is one of the main causes of death in critically ill patients. In recent years, with the emergence of new and drug-resistant pathogens and the gradual increase of immunosuppressed population, the incidence rate and mortality of infection remain high. Respiratory tract infection is one of the most common types of infection in clinical practice, typically characterized by acute onset, rapid progression, and complex pathogen composition. How to identify the pathogen in a short period of time is the key to early detection and treatment of respiratory infections. Recently, researchers have analyzed and mined more than 1000 clinical metagenome high-throughput sequencing diagnostic data generated by the local platform of zhongshan hospital. The results showed that alveolar lavage fluid was the best sample type for high-throughput sequencing detection of respiratory tract infection pathogens, and lung tissue and sputum could be selected as samples when mycobacterium tuberculosis infection was suspected; The host immune status is also one of the factors affecting the structure of respiratory microbiota, and the proportion of herpes virus detection is higher in immunocompromised hosts. Finally, the study once again confirmed that setting different bioinformatics diagnostic threshold standards for different pathogens can effectively identify pathogenic pathogens in respiratory samples, improve data utilization, and provide effective solutions for clinical automated analysis.
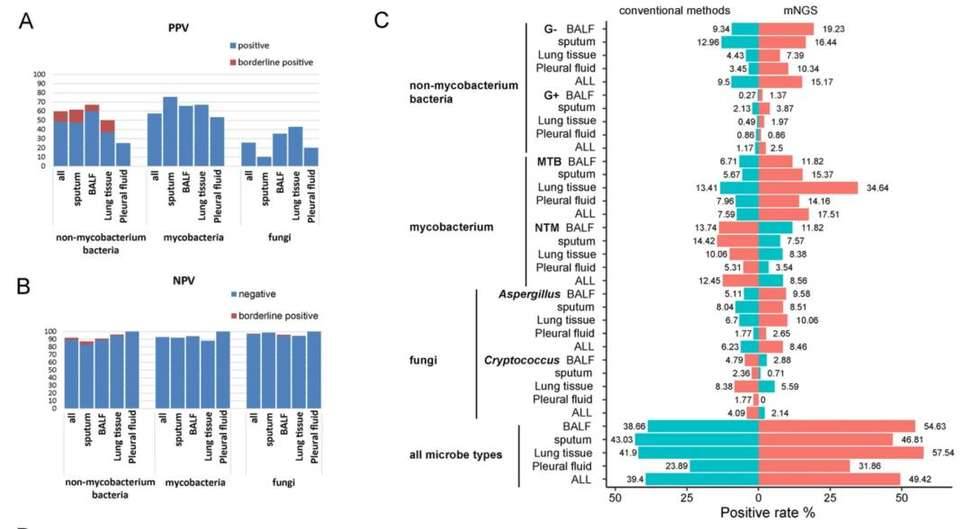 The detection performance of mNGS in different pathogens and samples, as well as the comparison of positive rates with traditional culture
The detection performance of mNGS in different pathogens and samples, as well as the comparison of positive rates with traditional culture
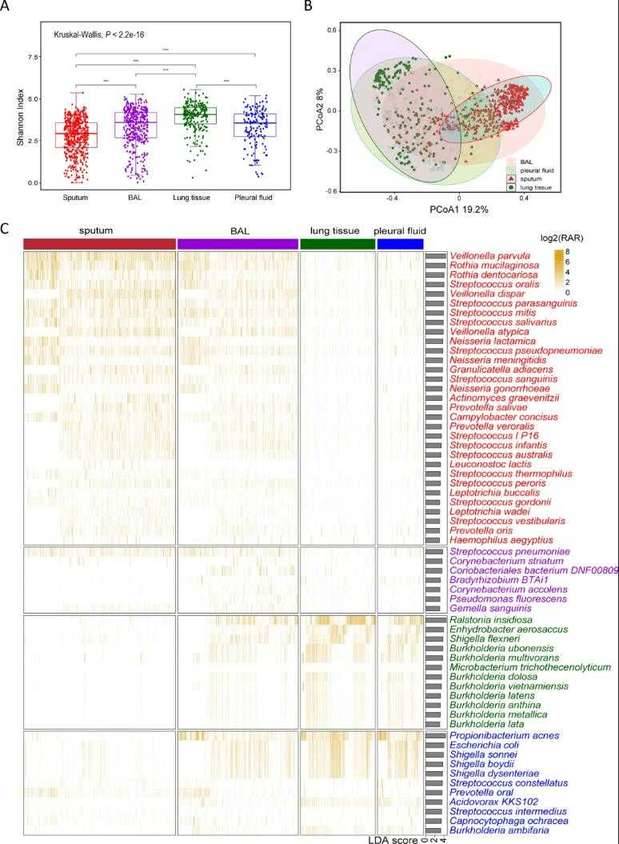 Microbial distribution in different patient populations
Microbial distribution in different patient populations
Metagenomics sequencing reveals pathogenic mechanisms of pathogens
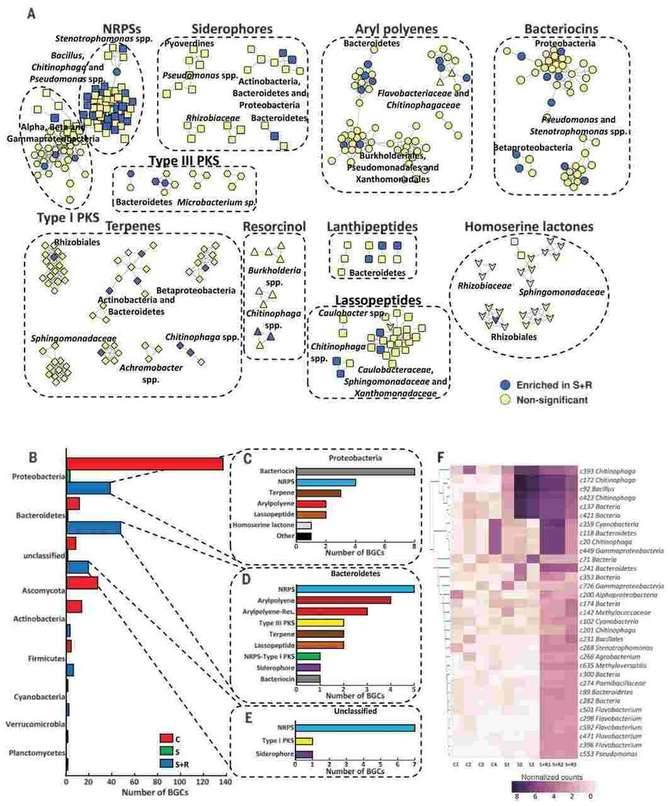
The cultivable microorganisms in plant endophytes are very limited. metagenomics is one of the effective means to study plant endophytes. Research on the metagenome of plant endophytes Through the research method of metagenomics that does not rely on pure culture, we construct a metagenome library of plant endophytes, and directly screen from this library to obtain biosynthetic gene cluster, further identify their functions, conduct qualitative and quantitative analysis of plant endophytes, and provide direct evidence for the origin of plant microorganisms. Some soils exhibit significant disease inhibition functions of plant pathogens, mainly attributed to the plant related microbiome. Microorganisms living inside plants can promote plant growth and health, but their genomic and functional diversity remain largely elusive. To understand the disease inhibitory function of endophytic bacteria living in plant root tissues, the author conducted metagenomic sequencing analysis on the roots of sugar beet seedlings grown in disease inhibiting soil. The microbial community and functional composition related to disease suppression were identified to distinguish which biosynthetic gene cluster (BGCs) were up-regulated during infection, and then the endophytic flora was recombined; Finally, perform site-specific mutagenesis analysis to determine whether specific BGCs play an important role in the disease suppression process. In the second stage of pathogen invasion into plant roots, endophytic bacteria can provide an additional protective layer. When pathogens invade, members of the Chitinophagaceae and Flavobacterium families accumulate in plants and exhibit enhanced enzyme activity related to fungal cell wall degradation, as well as biosynthesis of secondary metabolites encoded by NRPSs and PKSs. After reassembling 25 bacterial genomes using metagenomic sequences (de novo), a synthetic community (syncom) of flavobacterium and chitin bacteria can be reconstructed, providing disease protection. The results of this study highlight the abundant unknown microbial communities and functional traits in the endophytic microbiome. The category and function of microbial communities were successfully determined using metagenomic analysis and network inference, thereby obtaining plant phenotypes related to specific microbial groups.
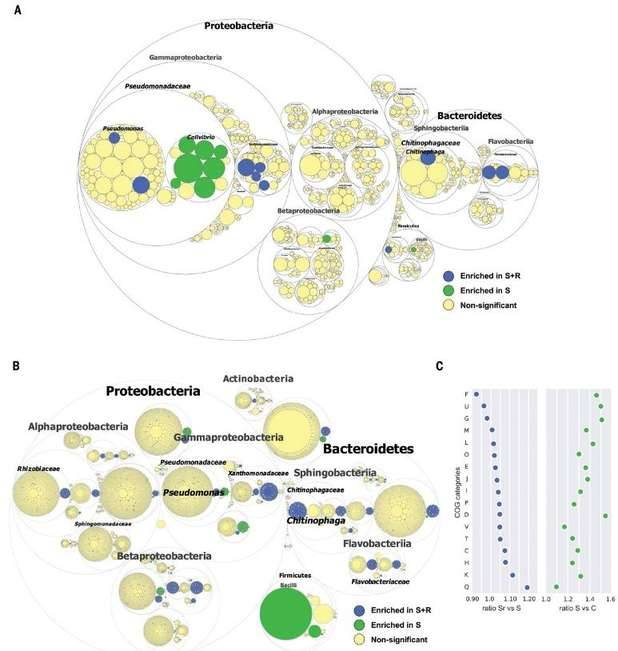 Pathogen-induced changes in endophytic microbiome diversity and functions.
Pathogen-induced changes in endophytic microbiome diversity and functions.
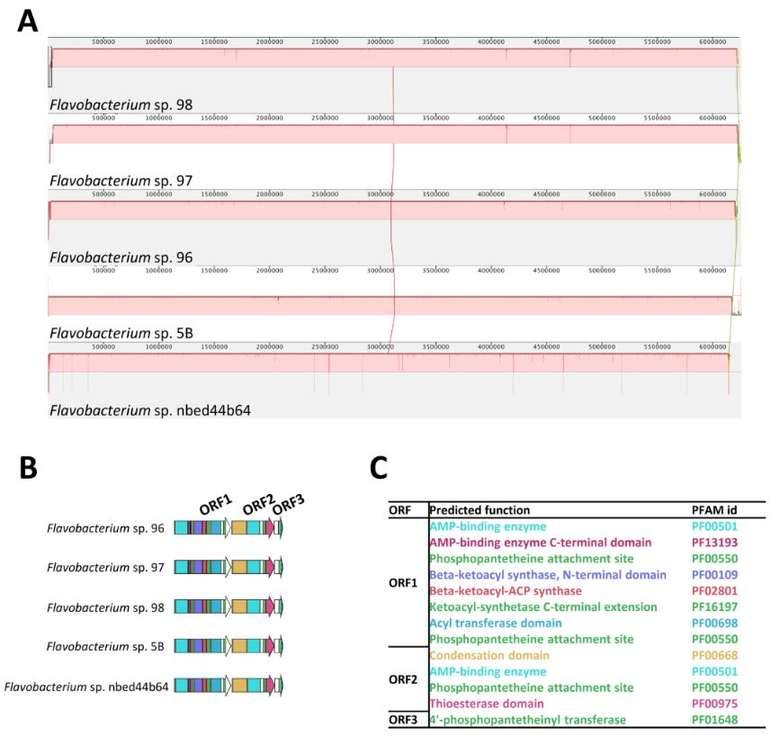 Diversity and distribution of biosynthetic gene clusters in the endophytic microbiome
Diversity and distribution of biosynthetic gene clusters in the endophytic microbiome
 Comparative Genomic Analysis of Sequenced Xanthobacterium Genomes and Macrogenomic Assembly Genomes (MAG)
Comparative Genomic Analysis of Sequenced Xanthobacterium Genomes and Macrogenomic Assembly Genomes (MAG)
Metagenomics, which uses the new generation of high-throughput sequencing (NGS) technology to study the microbial population genome in a specific environment, can further explore the functional activity of microbial population, mutual cooperation relationship and relationship with the environment, and explore potential biological significance on the basis of analyzing microbial diversity, population structure and evolutionary relationship.
References:
- Carrión, Víctor J et al. "Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome." Science (New York, N.Y.) vol. 366,6465 (2019): 606-612.
- Zeng, Qingchao et al. "Comparative genomic and functional analyses of four sequenced Bacillus cereus genomes reveal conservation of genes relevant to plant-growth-promoting traits." Scientific reports vol. 8,1 17009. 19 Nov. 2018.
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.


 Sample Submission Guidelines
Sample Submission Guidelines
 The detection performance of mNGS in different pathogens and samples, as well as the comparison of positive rates with traditional culture
The detection performance of mNGS in different pathogens and samples, as well as the comparison of positive rates with traditional culture Microbial distribution in different patient populations
Microbial distribution in different patient populations Pathogen-induced changes in endophytic microbiome diversity and functions.
Pathogen-induced changes in endophytic microbiome diversity and functions. Diversity and distribution of biosynthetic gene clusters in the endophytic microbiome
Diversity and distribution of biosynthetic gene clusters in the endophytic microbiome Comparative Genomic Analysis of Sequenced Xanthobacterium Genomes and Macrogenomic Assembly Genomes (MAG)
Comparative Genomic Analysis of Sequenced Xanthobacterium Genomes and Macrogenomic Assembly Genomes (MAG)