We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy


We are dedicated to providing outstanding customer service and being reachable at all times.
Improving the Accuracy of Nanopore Sequencing
At a glance:
- Overview
- Sources of Errors in Nanopore Sequencing Data
- Strategies to Improve the Accuracy of Nanopore Sequencing
Overview
Nanopore sequencing is a rapidly maturing length-based sequencing technology capable of analyzing natural DNA/RNA and sequencing fragments of any length, from short to ultra-long, making it possible to elucidate all types of genetic and epigenetic variation. However, while this technology has improved in recent years, it still exhibits relatively high error rates on raw sequences in the range of 5% to 15% when compared to standard next-generation sequencing (NGS) devices such as Illumina, and Oxford Nanopore continues to drive further performance enhancements by iteratively iterating on its technology to improve the accuracy of raw reads.
Over the years, Oxford Nanopore has iterated its technology to improve its performance, and they continue to improve the nanopore sensing system by updating analytical methods and new chemicals.
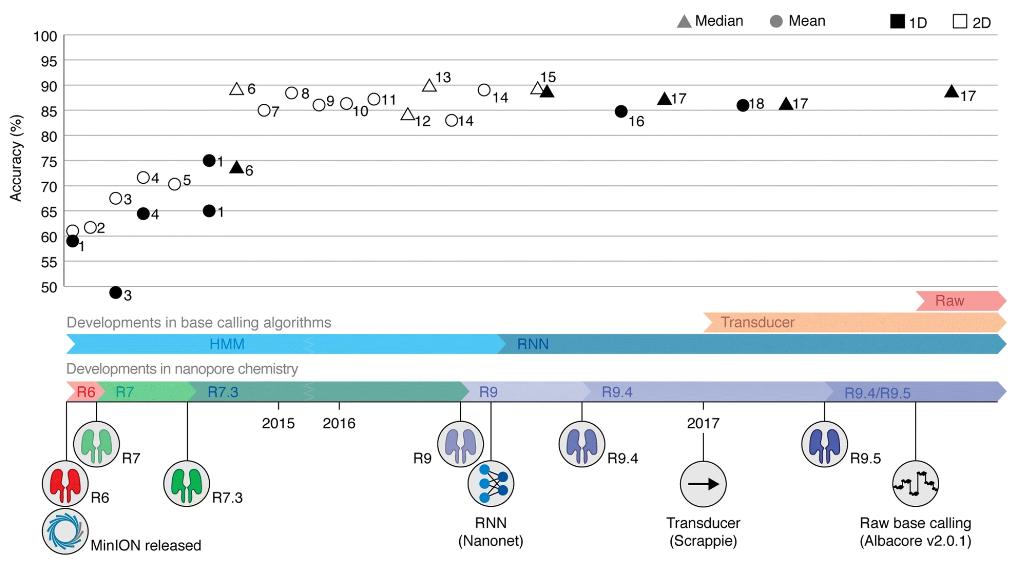 Timeline of reported MinION read accuracies and Oxford Nanopore Technologies (ONT) technological developments. (Rang F J et al., 2018)
Timeline of reported MinION read accuracies and Oxford Nanopore Technologies (ONT) technological developments. (Rang F J et al., 2018)
Sources of Errors in Nanopore Sequencing Data
Understanding errors in nanopore sequencing requires a comprehensive understanding of the mechanisms of the process. At the heart of a nanopore sequencing device are two electrolyte-filled pores separated by a membrane containing nanopores. When DNA is displaced through the nanopore, it causes interruptions in the ionic current. The nature and extent of these disruptions, caused by different nucleotides, form the basis for decoding DNA sequences.
Oxford Nanopore sequencing data may contain errors in the following two steps, which can be made more accurate with nanopore chemistry and basecalling software.
- Errors may occur during the sequencing process and therefore errors are inherent in the raw data. In this case, the inherent limitations of the technology result in a low signal-to-noise ratio, which prevents the identification of potential DNA sequences. There are several factors in the sequencing process that may contribute to a low signal-to-noise ratio:
(i) Structural similarity of nucleotides
(ii) The simultaneous effect of multiple nucleotides on the signal
(iii) The inhomogeneous rate of nucleotides passing through the pore
(iv) No change in signal within homopolymers - Errors can occur in the conversion of raw current signals into DNA sequences. Information about the DNA sequence is actually present in the data, but flaws in the analysis prevent its correct interpretation.
Strategies to Improve the Accuracy of Nanopore Sequencing
In any sequencing technology, accuracy is critical. For Nanopore sequencing, this is even more important given its unique method of directly reading nucleic acid sequences. Achieving high accuracy has multiple benefits, from ensuring correct variant calling to obtaining reliable shared sequences. It also extends the applicability of the technology in clinical diagnostics, environmental monitoring, and genetic research. To date, improvements in read accuracy can be achieved through several strategies:
Hole Optimization
Oxford Nanopore Technology (ONT) has made great strides in improving the nanopores themselves, the heart of its technology. Early versions (e.g., R6) were foundational but had their limitations. Moving into R9, this version heralded substantial enhancements. By improving the structure and function of the wells, ONT succeeded in improving the quality of the sequencing output. As the field progresses, we can anticipate the introduction of even more optimized wells to capture DNA or RNA sequences with even greater precision.
Raw Signal Processing
In contrast to segmented event data processing, raw signal processing has shown promise in improving sequencing accuracy. Tools such as BasecRAWller and Chiron utilize this approach. For example, Chiron utilizes a convolutional neural network to detect patterns in the raw data and then a recurrent neural network to predict k-mer probabilities. This approach eliminates errors that can occur during data segmentation, thus providing more accurate sequences.
Evolution of Library Preparation
One of the key factors determining the success of sequencing is how the DNA or RNA sample is prepared. ONT has innovated in this area by introducing methods such as 2D and 1D2 sequencing. These technologies allow multiple sequencing of a single DNA strand. By allowing multiple reads of the same fragment, the chances of capturing a more accurate sequence are multiplied.
Chemical Innovations
Novel chemicals can enhance nanopore sequencing accuracy. The introduction of R10.4.1 Flow Cell with Ligation Sequencing Kit V14 has achieved up to 99.5% raw read accuracy (Q23). This combination, when used in conjunction with duplex basecalling, has even exceeded 99.9% accuracy (Q30). These numbers are not just numbers but represent a huge leap forward in ensuring reliable genomic data.
Another noteworthy chemical advance is the ability to directly detect base modifications. Modifications to canonical bases (e.g., 5mC, 5hmC, and 6mA) have important epigenetic significance. Oxford Nanopore's technology is able to identify these modifications in real-time, without the need for additional sample preparation, marking a paradigm shift in epigenetic research.
Enhanced Basecalling Algorithms
At the heart of nanopore sequencing lies basecalling - the translation of raw electrical signals into nucleotide sequences. The earliest algorithms used Hidden Markov Models (HMM) to accomplish this task. Although effective, there is still room for improvement. The transition to recurrent neural networks (RNNs) has enhanced basecalling algorithms. RNNs, especially those using a bidirectional approach, can take into account information from greater distances and provide more accurate predictions. The transition from HMMs to RNNs has witnessed a huge leap in the quality of basecalling, resulting in more reliable sequencing results.
Utilizing Time-varying Cross Membrane Voltage
A groundbreaking approach to improving accuracy is the use of time-varying voltages in combination with DNA processing enzymes. Introduced in this study, this approach reduces two major sources of error in nanopore sequencing. By controlling DNA movement with different voltages, the researchers observed a significant increase in sequencing accuracy.
Consensus Calling and Refinement
Another method for improving accuracy is consensus calling. By comparing multiple nanopore reads and extracting shared sequences, random errors can be eliminated, leaving only systematic errors. Tools such as Nano Correct and Racon have been developed specifically for this purpose. In addition, post-sequencing correction tools like Nanopolish utilize raw data to improve (or refine) draft genome assemblies. The tool effectively utilizes the synthesized information in the raw signal (which is often overlooked in the final sequence) to fine-tune the assembly.
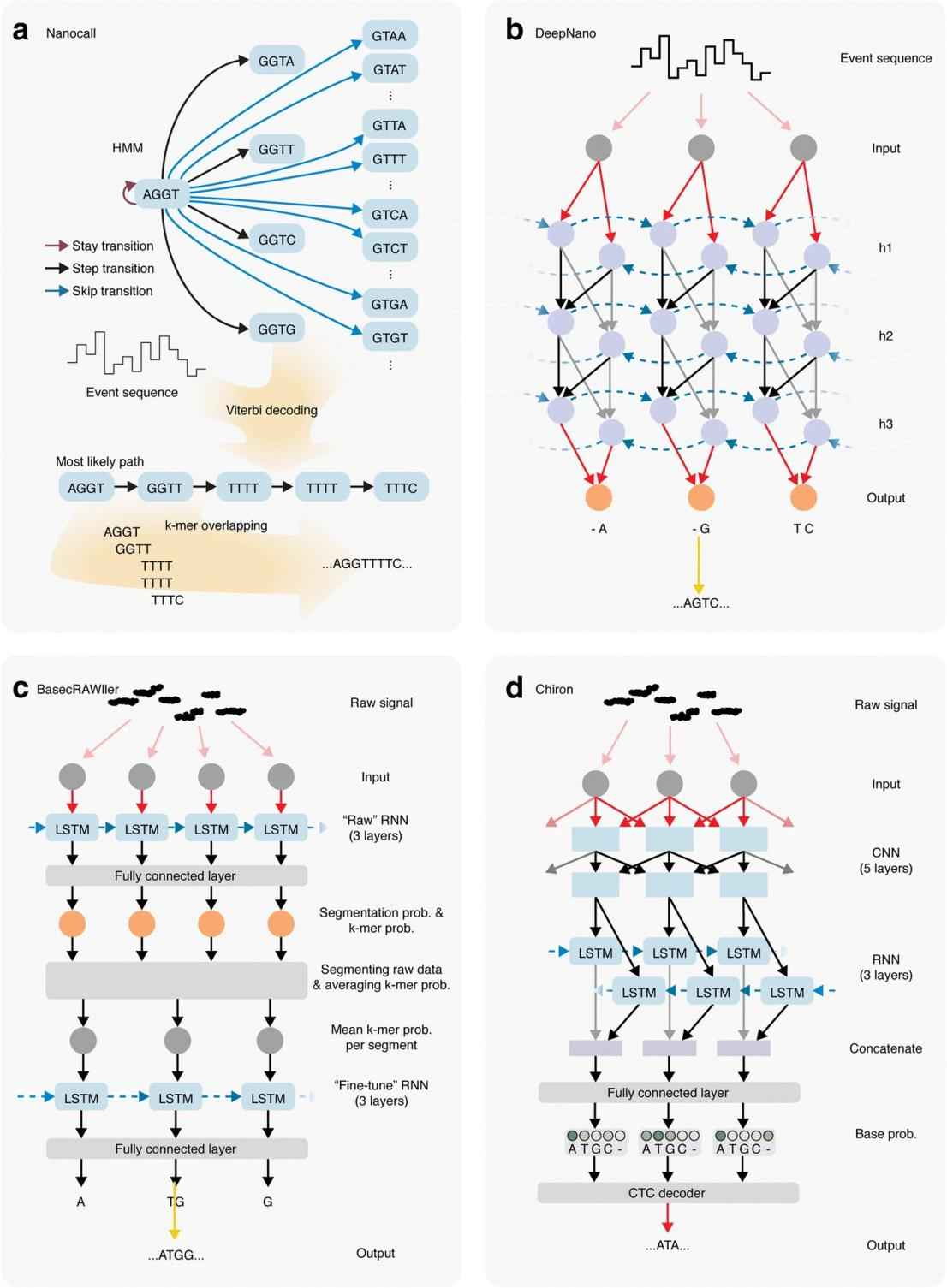 Schematic overview of the algorithms underlying nanopore base callers. (Rang F J et al., 2018)
Schematic overview of the algorithms underlying nanopore base callers. (Rang F J et al., 2018)
References
- Rang, Franka J., Wigard P. Kloosterman, and Jeroen de Ridder. "From squiggle to basepair: computational approaches for improving nanopore sequencing read accuracy." Genome biology 19.1 (2018): 90.
- Noakes, Matthew T., et al. "Increasing the accuracy of nanopore DNA sequencing using a time-varying cross membrane voltage." Nature biotechnology 37.6 (2019): 651-656.
Related Services
Oxford Nanopore Sequencing
Variant Calling
For research purposes only, not intended for personal diagnosis, clinical testing, or health assessment