We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy


We are dedicated to providing outstanding customer service and being reachable at all times.
Nanopore Basecalling
At a glance:
- Overview
- How Basecalling Works
- Neural Network-based Basecalling
- Nanopore Basecalling in the Perspective of Example Segmentation
Overview
Oxford Nanopore Technologies (ONT) represents the cutting edge of long-read sequencing technology, capable of generating long reads of tens of thousands of nucleotides. In addition to sequencing applications, it is used in microbiology, agriculture, and other fields. The primary function of this technology is to detect changes in ionic current signals as DNA or RNA fragments pass through a nanopore. Basecalling, the computational process of converting raw electrical signals into nucleotide sequences, is often the first step in analyzing nanopore sequencing signals and is critical for Nanopore sequencing platforms. As the technology matures, there is an urgent need to improve basecalling methods to ensure data reliability.
How Basecalling Works
When sequencing DNA or RNA using nanopores, the Oxford Nanopore sequencing device's MinKNOWTM software records the changes in electrical current caused by the passage of the DNA or RNA strand through the well. The nucleotides present in the pore affect the resistance of the pore, and the constant movement of bases through the pore results in a constant change in current (waveform curve.) MinKNOWTM processes the waveform graph in real time as reads, each corresponding to a DNA/RNA strand. These reads are written to a FAST5 file, which is the raw data collected by Nanopore. This raw data contains information not only about canonical bases, but also about base modifications, such as methylation.
Basecalling converts the raw current signals into nucleotide sequences and provides the nucleotide sequences for downstream analysis. This is not a simple task, as the electrical signals come from individual molecules, resulting in noisy and random data. ONT offers well-established software packages such as Scrappie and Guppy.
However, Nanopore basecalling still has a high error rate compared to short read-length sequencing. Their error rates range from 5% to 15%, while the Illumina Hiseq platform has an error rate of about 0.1%. Therefore, basecalling of ONT device signals becomes a challenging machine learning problem and a key determinant of ONT sequencing quality and usability.
In this work, the researchers examined the performance of different basecalling tools, looking at the accuracy of base levels in individual reads as well as the accuracy of majority rule consensus basecalling in the assembly. They also looked at a number of other aspects of basecalling: training with datasets specific to classification units, using larger neural network models, and improving consensus basecalling in assemblies by performing additional signal level analysis using Nanopolish.
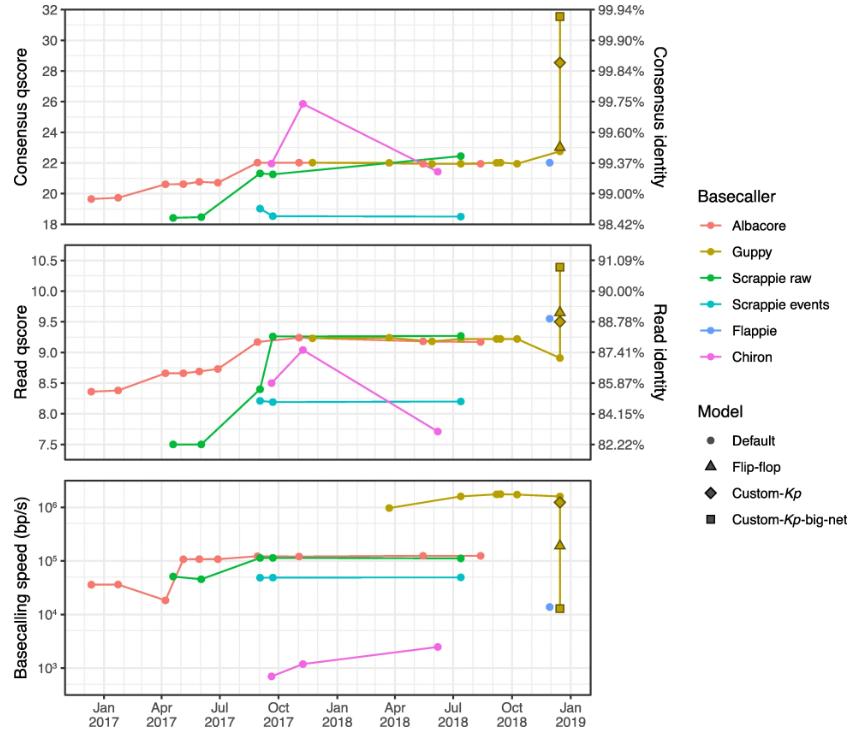 Read accuracy, consensus accuracy and speed performance for each basecaller version, plotted against the release date. (Wick R R et al., 2019)
Read accuracy, consensus accuracy and speed performance for each basecaller version, plotted against the release date. (Wick R R et al., 2019)
Neural Network-based Basecalling
Basecalling is an active field, with both ONT and independent researchers developing methods.The basecalling algorithms currently deployed by ONT are based on neural networks. They loosely mimic biological neural networks within the human brain, with connections of "nodes" (equivalent to neurons) passing data between them to reach a predicted sequence of bases. Crucially, just like the human brain, these neural networks have the ability to learn and improve their predictions over time.
Guppy currently uses bidirectional recurrent neural networks where information can be passed back and forth between nodes. Alternative algorithms are continually evaluated for their applicability to basecalling, such as convolutional neural networks, which are commonly used for image and video processing because the connections between nodes mimic the organization of the brain's visual cortex.
However, such algorithms must be trained using real data. As a result, the performance of any given base recognizer is affected by the data used to train its model, especially when basecalling is performed on natural (non-PCR amplified) DNA containing base modifications.
Nanopore Basecalling in the Perspective of Example Segmentation
Traditionally, basecalling has been viewed primarily as a sequence tagging challenge. However, new approaches have emerged that address the problem from different perspectives. One of the avant-garde perspectives is instance segmentation. Base detection is not just about sequence tagging, but can also be envisioned as a multi-label segmentation task. The approach aims at segmenting the original signal and assigning the corresponding nucleotide labels.
The UR-net model is an evolution of the basic U-net model and is specifically designed to manage one-dimensional segmentation tasks, capturing the sequential complexity in base identification tasks. This approach allows direct processing of raw signals without having to segment them first. Examining basecalling through this lens has the potential to improve accuracy by integrating basecalling and segmentation.
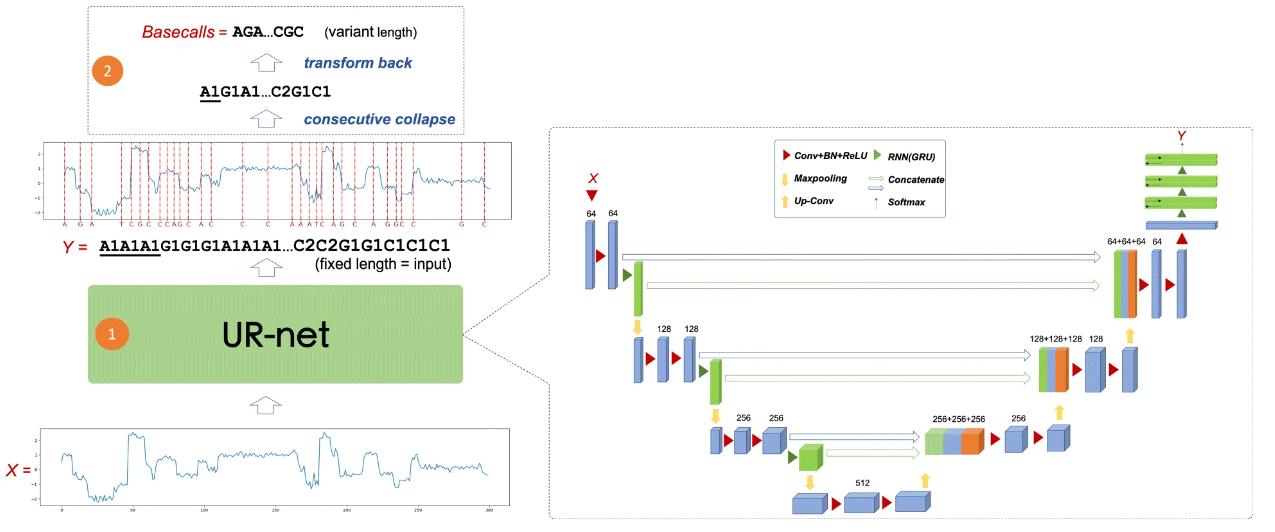 Overall pipeline of URnano basecaller. (Zhang Y et al., 2020)
Overall pipeline of URnano basecaller. (Zhang Y et al., 2020)
References
- Wick, Ryan R., Louise M. Judd, and Kathryn E. Holt. "Performance of neural network basecalling tools for Oxford Nanopore sequencing." Genome biology 20 (2019): 1-10.
- Zhang, Yao-zhong, et al. "Nanopore basecalling from a perspective of instance segmentation." BMC bioinformatics 21 (2020): 1-9.
Related Services
Oxford Nanopore Sequencing
Long-Read Sequencing of DNA Methylation
Long-Read Sequencing of RNA Methylation
For research purposes only, not intended for personal diagnosis, clinical testing, or health assessment