We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy


We are dedicated to providing outstanding customer service and being reachable at all times.
Long Read Sequencing for Infectious Disease Research
Infectious diseases, which are caused by organisms such as bacteria, viruses, fungi, or parasites, remain a major threat in today's world. The fight against infectious diseases begins with prevention, diagnosis, and treatment. Diagnosis can be maintained by looking at the causative agent under a microscope or by detecting the presence of nucleic acids and proteins of the pathogen. Molecular techniques range from classical polymerase chain reaction (PCR) to sequencing of nucleic acid components. CD Genomics is a leading global life sciences company providing specialized long read sequencing solutions for infectious disease research. Our goal is to help our customers detect pathogens, analyze antimicrobial resistance (AMR), and characterize high-quality genome assembly and variation.
Introduction to Infectious Diseases
Infectious diseases are diseases caused by harmful organisms (pathogens) that enter the body from the outside. The pathogens that cause infectious diseases are viruses, bacteria, fungi, parasites, and rarely prions. You can get infectious diseases from other people, mosquito bites, and contaminated food, water, or soil. Initially, it was thought that such diseases would be eradicated quickly, but adaptation of pathogens to environmental stresses, such as antimicrobial drugs, has facilitated the emergence and re-emergence of infectious diseases. The definitive diagnosis of many infectious diseases still relies on direct observation of the causative pathogen. However, many times this is difficult to achieve, especially for viral diseases, because of the size of the viral particles. As a result, molecular techniques such as PCR or enzyme-linked immunosorbent assays have been developed, which allow direct measurement of nucleic acids or proteins of infecting pathogens to aid in more reliable diagnosis. However, these techniques cannot diagnose or test for microbial drug resistance. Long read sequencing has emerged as an attractive option for in situ diagnosis of infectious pathogens, facilitating a potentially rapid response to the identification and management of disease sources and disease transmission.
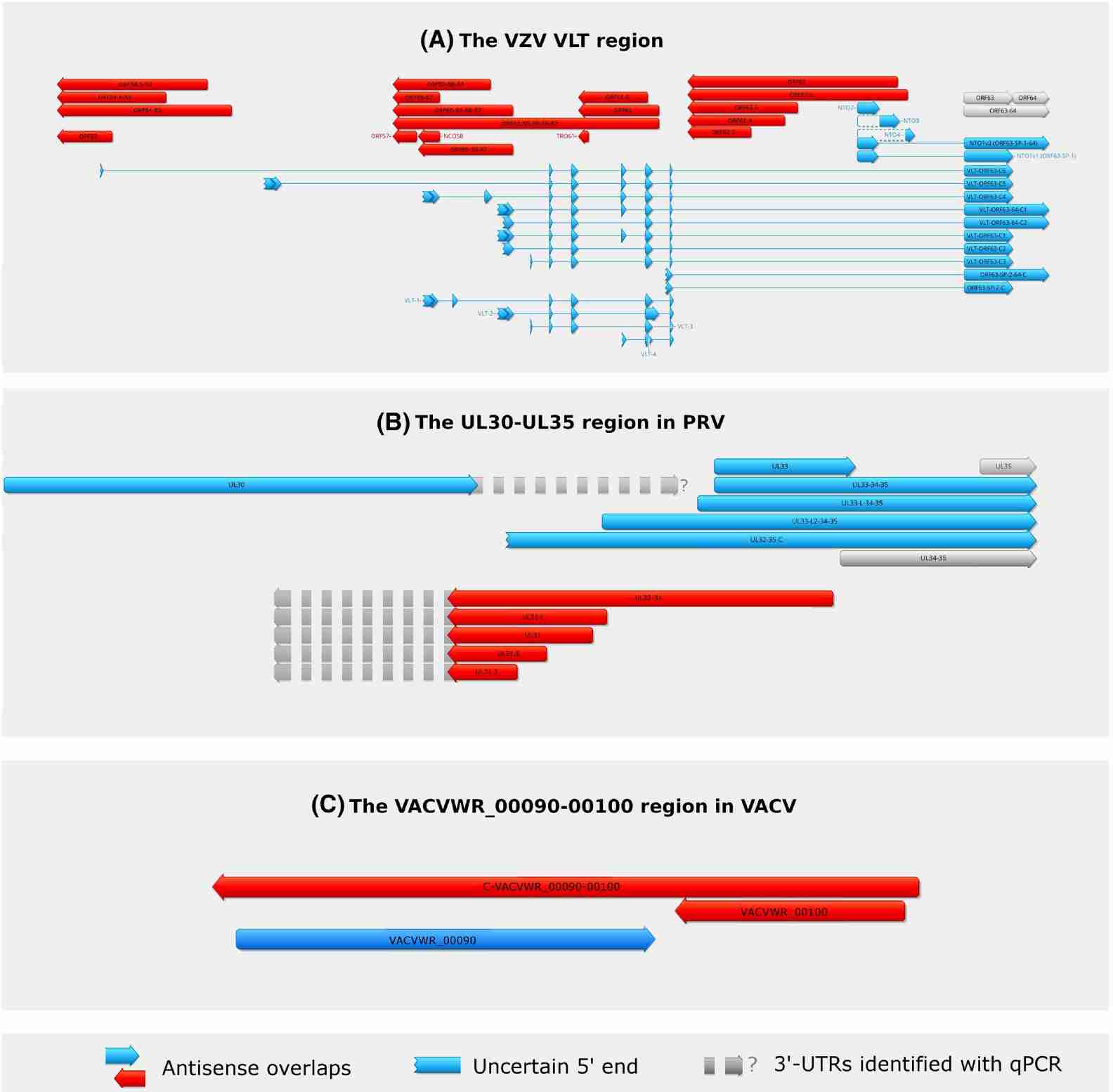 Fig. 1. Antisense Transcripts in Two Herpesviruses. (Boldogkői et al., 2019)
Fig. 1. Antisense Transcripts in Two Herpesviruses. (Boldogkői et al., 2019)
Solutions Offering at CD Genomics
Based on our advanced PacBio SMRT sequencing and ONT Nanopore sequencing platforms, sequencing is performed in the lab or at the sample source at a scale to meet your needs. We aim to provide comprehensive genetic information to increase our customers' understanding of microbial genomics, pathogenicity, and antimicrobial resistance.
Our long read sequencing solutions are tailored for infectious disease research, providing comprehensive, real-time insight into infectious disease samples, including pathogen identification as well as characterization of virulence factors, strain typing, and antibiotic resistance markers. Our long read sequencing services for infectious disease research are as follows.
- Pathogen Detection
We offer long read sequencing for the rapid detection of pathogens, including the diagnosis of bacterial meningitis, bacterial lower respiratory tract infections, infective endocarditis, pneumonia, prosthetic joint infections, sexually transmitted infections, etc. Our solutions enable real-time genomic monitoring of epidemics, helping to elucidate patterns of viral evolution, monitor the efficacy of diagnostic assays, and study the chain of transmission.
- Antimicrobial Resistance (AMR) Analysis
Our long read sequencing can also analyze antibiotic/antimicrobial drug resistance in bacteria and other microorganisms. Our solutions allow comprehensive and accurate characterization of microbial genomes, as well as the study of antibiotic resistance mechanisms and the identification of new resistance genes. Genotypic resistance testing based on long read sequencing is widely used for a variety of viral infections, including HIV-1, cytomegalovirus (CMV), hepatitis B, hepatitis C, and influenza.
We offer a simple workflow for high-resolution and rapid differentiation of intestinal, vaginal, and other microbiota by long read sequencing. The process combines long-read sequencing with minimally invasive sampling procedures for intestinal and vaginal specimens with low bacterial loads. The analysis has a very short turnaround time of 4 hours and is economically feasible. It allows accurate estimation of microbiome composition compared to traditional studies that rely on 16S rRNA and DNA amplicons.
Applications of Our Long Read Sequencing Solutions for Infectious Disease Research
- Generate closed microbial genomes and plasmids to reveal drivers of pathogenicity, virulence, and drug resistance.
- Resolve viral populations to reveal host-pathogen dynamics and advance drug and vaccine design and evaluation.
- Characterize microbial populations to understand how the microbiome impacts human health.
Advantages of Our Long Read Sequencing Solutions for Infectious Disease Research
- Complete sequencing of genes to improve resolution for detecting infectious bacterial species.
- Multiple bioinformatics tools combine to improve amplicon accuracy and determine bacterial composition up to the species level.
- Pathogens can be identified within 2h of sequencing and a definitive diagnosis can be obtained in less than 4h.
- Direct detection of DNA/RNA methylation.
- Whole-genome, metagenome, targeted, direct RNA, and cDNA sequencing methods.
- Uses ultra-long read sequencing protocols, with special protocols to obtain read lengths of >2 Mb.
CD Genomics is committed to providing accurate, comprehensive, and high-resolution long read sequencing solutions to accelerate infectious disease research. Our goal is to utilize long-read sequencing to support rapid identification and control of infectious disease outbreaks. If you have any questions, please feel free to contact us. We look forward to working with you on projects of interest.
Reference
- Boldogkői, Zsolt, et al. "Long-read sequencing–a powerful tool in viral transcriptome research." Trends in microbiology. 27.7 (2019): 578-592.
Related Services
PacBio SMRT Sequencing
Oxford Nanopore Sequencing
Microbial Genomics with Long-Read Sequencing
Antibiotic Resistance Gene (ARG) Analysis
Full-Length Plasmid Sequencing
Full-Length 16S/18S/ITS Amplicon Sequencing
For research purposes only, not intended for personal diagnosis, clinical testing, or health assessment