We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy


We are dedicated to providing outstanding customer service and being reachable at all times.
Antibiotic Resistance Gene (ARG) Analysis

Nowadays, antibiotics have become a powerful weapon in the prevention and treatment of bacterial infections. With this comes the growing problem of antibiotic resistance. The spread of antibiotic resistance has weakened the effectiveness of antibiotics, resulting in more than 700,000 deaths per year and becoming a global public health threat. Here, CD Genomics is providing antibiotic resistance gene (ARG) analysis services using long-read (ONT Nanopore) sequencing. This advanced third-generation sequencing technology can generate large amounts of long sequences in a fast and cost-effective manner and has been successfully exploited to detect antimicrobial ARGs. With the help of our services, customers can detect and identify ARGs, involving assessing their taxonomic origin, genetic organization, bacterial hosts, and so on.
Overview of Antibiotic Resistance Gene (ARG) Analysis
Since antibiotic resistance is one of the most global public health crises, it is essential to understand the mobility risk of ARGs in the environment and the pathways of their transmission. ARGs are diverse and abundant, occurring widely in soil, sewage, aquatic ecosystems, and animal manure. In particular, due to the wide use of antibiotics in the animal farming industry, ARGs in livestock feces are noticeable. The prolonged presence of antibiotics in animal feces increases the emergence of functional antibiotic resistance and the risk of horizontal gene transfer (HGT). In addition to exerting selection on microbes, a large number of human commensals in animal feces increases the potential for ARG transfer to human-associated pathogens. The determination and study of ARGs in environmental systems have benefited from the next-generation sequencing (NGS). High-throughput sequencing (such as Illumina sequencing) capability overcomes the limitations of PCR-based methods with limited detection capacity and component bias. Despite the cost-effectiveness and low error rate of NGS technology, its short read length makes the assembly and analysis of contiguous genomic sequence regions containing multiple co-localized ARGs and/or mobile genetic elements (MGEs) challenging. In contrast, long reads generated by ONT Nanopore sequencing can reveal co-localized ARGs as well as their flanking genomic nucleotide sequences, providing additional information for better exploration of ARGs.
Advantages of Long-Read Sequencing in ARG Analysis
- The ability to reveal gene location, host range, and co-selection pattern of a resistome.
- The ability to assemble and identify more plasmids.
- The ability to identify functional complexes and operons.
Workflow of Our ARG Analysis Service

Sample Requirements
- Sample Type: Genomic DNA, OD260/280=1.8~2.0, no degradation and no contamination.
- DNA amount: ≥ 1μg.
- Sequencing strategy: Nanopore PromethION platform.
Analysis Pipeline
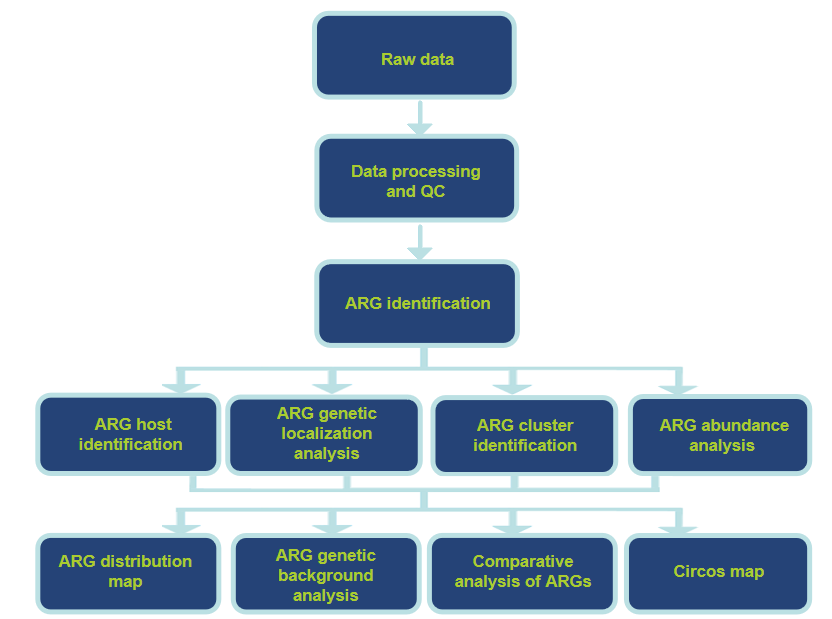
Our Service Support
- The profiling of ARGs and their neighboring genes.
- Detection and identification of ARGs, including their abundance, genetic locations, and mobility potential.
- Identification of ARG cluster.
- Species information annotated by ARGs.
- Comparative analysis of ARGs under different contexts.
CD Genomics can greatly improve our customer's understanding of the antibiotic resistome and their microbial contexts in environmental samples. Notably, our service is almost applicable to most types of environmental samples. Let us know your research objectives and our team of experts will contact you within one business day to discuss your needs.
References
- Qian, X., et al. (2021). "Long-read sequencing revealed cooccurrence, host range, and potential mobility of antibiotic resistome in cow feces." Proceedings of the National Academy of Sciences, 118(25).
- Dai, D., et al. (2022). "Long-read metagenomic sequencing reveals shifts in associations of antibiotic resistance genes with mobile genetic elements from sewage to activated sludge." Microbiome, 10(1), 1-16.
For research purposes only, not intended for personal diagnosis, clinical testing, or health assessment
