1 Introduction to lncRNA
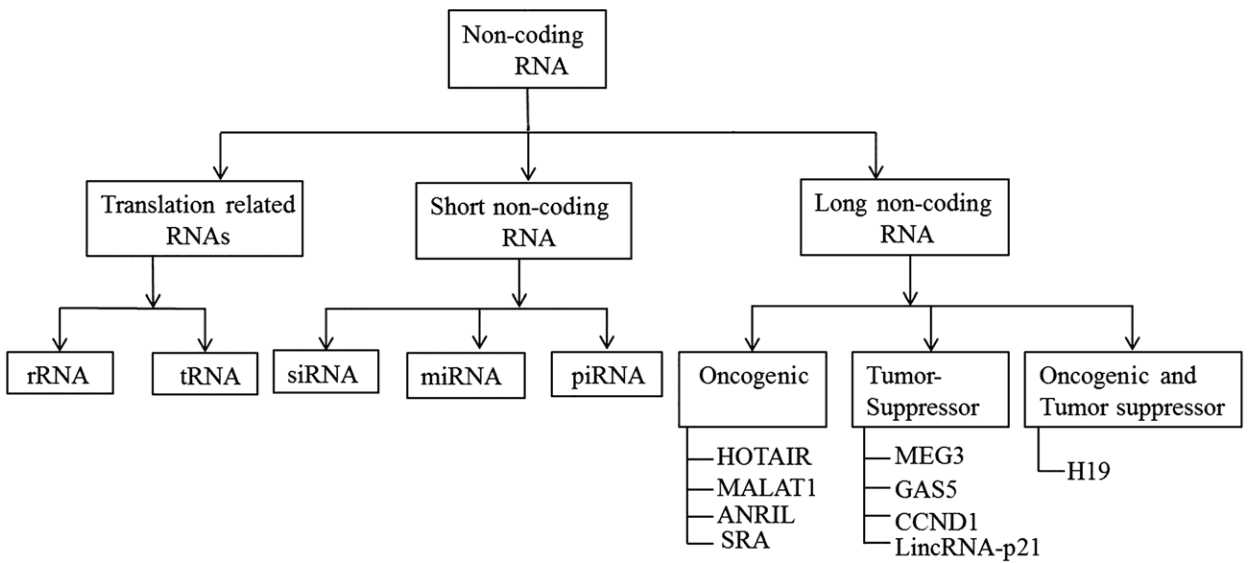
Non-coding RNAs (ncRNAs) are considered non-translated transcripts of genomic sequences. The classification of ncRNAs based on their functions is presented in Figure 1A. Long non-coding RNAs (lncRNAs) are a type of ncRNAs that are longer than 200 nt. They frequently reside in the nucleus and are tissue-specific. Compared to protein-coding mRNAs, lncRNAs have poorer interspecies sequence conservation. According to lncRNA position, lncRNAs are roughly divided into two types:
- Intergenic lncRNAs, including pseudogenes, long intergenic ncRNAs (lincRNAs), and very long intergenic ncRNAs (vlincRNAs/macroRNAs)
- Gene-overlapping lncRNAs, represented by intronic RNAs, natural antisense transcripts (NATs), promoter RNAs, and others
There are multiple theories about lncRNA origins and evolution, such as gene duplication and generation by mobile genetic element insertion. Extensive efforts have been devoted to a better understanding of lncRNA biology. And multiple databases of functional lncRNA information have been established, such as lncRNAbd, NONCODE, LncRNADisease, lncRNAtor, ncFANs, lnCeDB, LNCipedia. Table 1 exhibits the common lncRNA informatics resources.
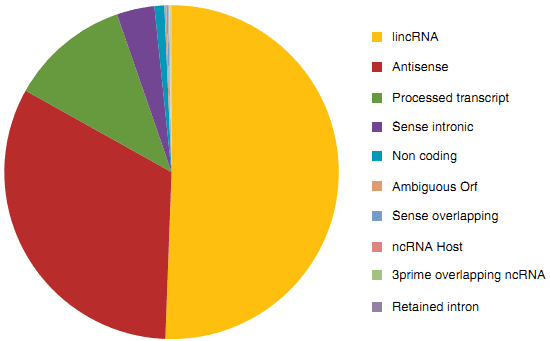
Figure 1. A subset of ncRNAs and their proportions of the noncoding transcriptome in human.
The application of next-generation sequencing technology has greatly facilitated the discovery and function analysis of lncRNAs. LncRNA sequencing is a powerful tool to profile whole-genome lncRNAs at single-base resolution, providing insights into their distribution, function, regulatory network, and disease implications. You can view this article Workflow of LncRNA Sequencing and Its Data Analysis for more details about lncRNA sequencing.
Table 1. The lncRNA bioinformatics resources.
| Name |
Description |
Website |
| NRED |
Database of lncRNA expression |
http://nred.matticklab.com/cgi-bin/ncrnadb.pl/ |
| ncFANs |
Web server for functional annotation of lncRNAs. |
http://ebiomed.org/ncfans/ |
| lncRNAdb |
Database of comprehensive annotations of functional lncRNAs. |
http://www.lncrnadb.org/ |
| NONCODE |
Database of integrative annotations of lncRNAs. a |
http://www.noncode.org/NONCODERv3/guide.htm |
| LNCipedia |
Database of annotations and structures of lncRNA sequences. |
http://www.lncipedia.org/ |
| LncRNADisease |
Database of lncRNA-associated diseases. |
http://cmbi.bjmu.edu.cn/lncrnadisease/ |
| DIANA-LncBase |
Database of microRNA targets on lncRNAs. |
http://www.microrna.gr/LncBase/ |
| iSeeRNA |
The lincRNA transcripts identified from transcriptome sequencing data. |
http://www.myogenesisdb.org/iSeeRNA/ |
| ChIPBase |
Database for annotating and exploring the expression profiles in transcriptional regulation of lncRNAs and other ncRNAs. |
http://deepbase.sysu.edu.cn/chipbase/ |
| lncRNAtor |
LncRNA portal encompassing expression profile, interacting (binding) protein, integrated sequence curation, evolutionary scores, and coding potential. |
http://lncrnator.ewha.ac.kr/index.htm |
| lnCeDB |
database of human lncRNAs that can potentially act as competing endogenous RNAs (ceRNAs). |
http://gyanxet-beta.com/lncedb/ |
2 Disease-specific applications of lncRNAs
Emerging researches suggest that lncRNAs play an important role in a wide range of biological processes, as well as in the pathophysiology of disease processes. And deregulated lncRNA expression has vital consequences during malignant transformation. In addition to cancer, lncRNAs are found to have roles in neurodegenerative disorders (such as Huntington’s disease and Alzheimer’s disease), brain development, cardiovascular disease, diabetes, and other common and rare diseases.
2.1 LncRNAs as biomarkers
Exosomes are considered a highly promising source of biomarkers in cancer. LncRNAs have been identified in exosomes, and have also been implicated in TGFβ dependent chemoresistance pathways. HOTAIR is a lncRNA and has been implicated in the pathogenesis of many malignancies. For example, HOTAIR can suppress PRC-2-dependent gene and its over-expression is a powerful predictor of metastasis and death. Exosomal HOTAIR expression can be used for distinguishing benign from malignant disease with a specificity of 57.2% and a sensitivity of 92.3%. But in combination with miR-21, the specificity and sensitivity increase to 73.5% and 94.2%, respectively.
In addition to HOTAIR, there are many other lncRNAs that are promising disease biomarkers. LncRNA MALAT1 is a tumor marker that is over-expressed in multiple types of tumor. HULC was found to be significantly over-expressed in live cancer, metastatic hepatic colorectal cancer, and HBV. PCGEM1 was detected in prostate tumors to be correlated with increased proliferation and colony formation. PCNCR1 is another described prostate cancer biomarkers, which was identified on chromosome 8q24.2. PCNCR1 is expressed as a 13 kb transcript and can trans-activate androgen receptor (AR), an important player in the progression of prostate cancer. DD3 was also found highly up-regulated in prostate cancer but its role in prostate cancer progression is still unknown.
2.2 LncRNAs-targeted therapeutic agents
LncRNAs may be used as therapeutic agents with the help of gene therapy delivery systems such as viruses. On the other hand, lncRNAs can be targeted with synthetic siRNAs or miRNAs, or drugs that specifically interact with lncRNAs to achieve therapeutic effects. There are two significant advantages of lncRNA-targeted drugs. One is that they allow protein upregulation/activation which was previously unsuccessful with small molecules drugs. The other is that lncRNA-targeted drugs have a high specificity, which would be beneficial especially in case when target proteins have multiple closely related family members, such as ion channels and protein kinases.
There are primarily two classes of oligonucleotide-based approaches: (i) antisense-based and (ii) catalytic oligonucleotide-based methods. The former method has been widely used to decipher functions of lncRNAs or as a therapeutic strategy. The latter method includes both DNAzymes and RNAzymes. Catalytic oligonucleotides can also be used to understand the functions of ncRNAs. Small molecules may have specific effects on modulating lncRNA activity. Such interest and endeavor as unbiased high-throughput screening, computational modeling of RNA structures, and design of novel scaffolds with specificity to RNA ligand have greatly accelerated small molecule-based targeting of lncRNA.
Conclusions
Although significant progress has made in the understanding of the biology of lncRNA and related therapeutic applications, the lncRNA field is still in its infancy. The future lncRNA researches may focus on the regulation of lncRNA expression, their isoform composition and developmental- and tissue-specificity.
At CD Genomics, we can provide reliable lncRNA sequencing service and professional bioinformatics analysis. Other non-coding RNA sequencing services are also available, including small RNA sequencing, circRNA sequencing, and degradome sequencing. Please feel free to contact our scientist for detailed service specifications.
References:
- Bhartiya D, Kapoor S, Jalali S, et al. Conceptual approaches for lncRNA drug discovery and future strategies. Expert opinion on drug discovery, 2012, 7(6): 503-513.
- Khorkova O, Hsiao J, Wahlestedt C. Basic biology and therapeutic implications of lncRNA[J]. Adv Drug Deliv Rev, 2015, 87:15-24.
- Santosh B, Varshney A, Yadava P K. Non-coding RNAs: biological functions and applications. Cell biochemistry and function, 2015, 33(1): 14-22.
- Spizzo R, Almeida M I, Colombatti A, et al. Long non-coding RNAs and cancer: a new frontier of translational research?. Oncogene, 2012, 31(43):4577-87.
- Tang J Y, Lee J C, Chang Y T, et al. Long Noncoding RNAs-Related Diseases, Cancers, and Drugs. The Scientific World Journal,2013,(2013-6-6), 2013, 2013(2):943539.
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.


 Sample Submission Guidelines
Sample Submission Guidelines