In recent years, long chain non-coding RNA (LncRNA) has become a research hotspot in the scientific research field and extensive studies have revealed multifaceted roles of long noncoding RNAs (lncRNAs) in gene regulation, accompanying an increased understanding of lncRNA processing, localization, interacting macromolecules and structural modules. Emerging evidences have recently suggested the crucial role of long noncoding RNAs (lncRNAs) in the tumorigenesis and progression. Various lncRNAs have been shown to possess aberrant expression and participate in cancerous phenotypes through their binding with DNA, RNA or proteins, or encoding small peptides.
Exploring the molecular mechanism of colorectal cancer metastasis through LncRNA
Long non-coding RNA (lncRNA) is a new major regulator of cell invasion and metastasis and the long non-coding RNAs (lncRNAs) are a new master regulator of cell invasion and metastasis. The survival rate of patients with liver metastasis from colorectal cancer is still very low. The number of CRLM regulators and clinical indicators remains limited. However, the number of CRLM regulatory factors and clinical indicators is still limited. CRLM1 was weakly associated with the chromatin regions of genes involved in cell adhesion and DNA damage, and this association was bidirectionally correlated with CRLM1-regulated pro-metastatic gene expression. This research investigated the expression dynamics of lncRNA in CRLM, primary CRC, and normal tissues, and discovered a series of lncRNAs related to metastasis, including CRLM1. CRLM1 inhibits apoptosis of Balb/C nude mouse CRC cells and promotes liver metastasis. CRLM1 is weakly related to the chromatin region of genes involved in cell adhesion and DNA damage, and this association is bi-directional related to the expression of pre transferred genes regulated by CRLM1. CRLM1 interacts physically with hnRNPK protein and promotes its nuclear localization. CRLM1 effectively enhanced the occupancy rate of hnRNPK promoter and co regulated the expression of a group of transfer genes. This study explored the molecular mechanism of CRLM1 through RNA sequencing (RNA seq), RNA purified chromatin separation (ChiRP), immunofluorescence (IF) and other methods. The results showed that there were CRLM related long non-coding RNA. CRLM1, one of these lncRNAs, not only predicted a poor prognosis, but also inhibited apoptosis and promoted metastasis in vivo and in vitro. These results help us understand how lncRNA regulates transcription through trans-regulation and identify prognostic markers and therapeutic targets for CRLM.
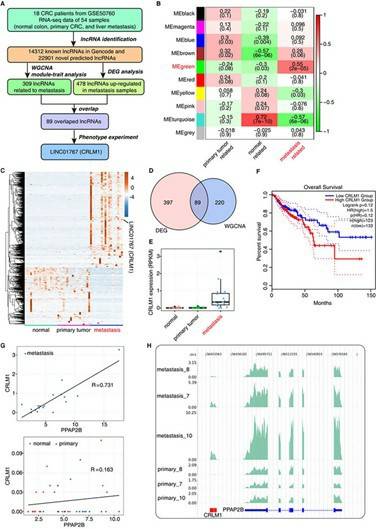 Overexpression of CRLM1 in colorectal liver metastases and associated with low survival rate
Overexpression of CRLM1 in colorectal liver metastases and associated with low survival rate
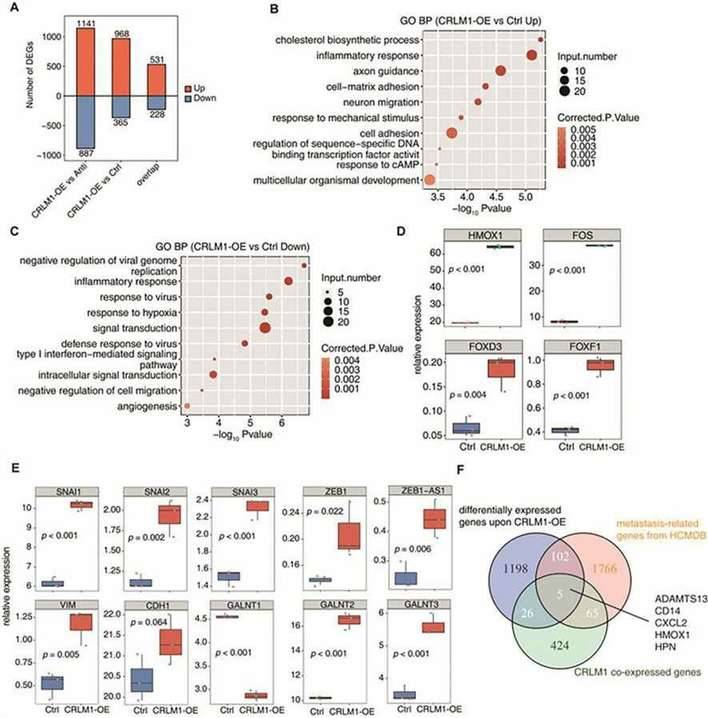 CRLM1-OE converts CRC cell transcriptome into metastatic state
CRLM1-OE converts CRC cell transcriptome into metastatic state
Understanding the biological effects and functional mechanisms of lncRNA promotes the progression of liver cancer
Hepatocellular carcinoma (HCC) is one of the most common malignancies worldwide and ranks as the third most common cause of cancer-related death. Researchers performed RNA seq on clinical liver cancer tissue and paired adjacent normal liver tissue to identify differentially expressed lncRNAs. By crossing the WCH and TCGA queues, 101 significantly altered lncRNAs were identified. Subsequently, based on the survival analysis of the TCGA LIHC queue, 5 candidate lncRNAs were successfully identified. Next, the expression levels of these candidate lncRNAs were verified using qRT PCR on 20 pairs of tissue samples and TLNC1 was identified as a potential tumorigenic lncRNA of liver cancer. TLNC1 significantly enhanced the growth and metastasis of hepatoma cells both in vitro and in vivo. In summary, this study indicates that TLNC1 can promote tumor growth and metastasis of liver cancer in vitro and in vivo. In terms of mechanism, we have demonstrated that TLNC1 can interact with TPR, strengthen the interaction between TPR and CRM1, and promote the nuclear output of p53, leading to downregulation of a series of tumor suppressors and upregulation of many oncogenes, ultimately contributing to the induction of liver cancer growth and metastasis. Additionally, TLNC1 is a promising prognostic factor of liver cancer, and the TLNC1-TPR-p53 axis can serve as a potential therapeutic target for hepatoma treatment.
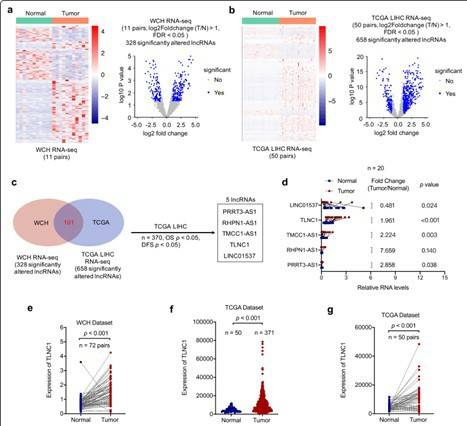 TLNC1 is upregulated in liver cancer tissues
TLNC1 is upregulated in liver cancer tissues
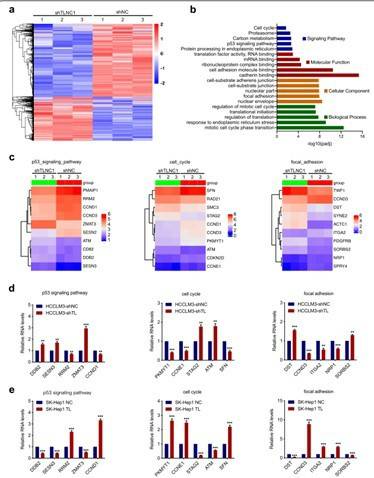 TLNC1 interacts with TPR and represses the transcriptional activity of p53
TLNC1 interacts with TPR and represses the transcriptional activity of p53
LncRNA is gradually becoming one of the hot topics in RNA biology and oncology research. Understanding how lncRNA-macromolecule interactions occur is key for dissecting their regulatory mechanisms. Without doubt, the multiple facets of lncRNA-mediated gene regulation have added a new dimension to the central dogma of gene regulation. Challenges remain at multiple levels, however, to fully dissect their functions. Characterizing lncRNAs and their isofoms in a spatial-temporal manner in normal, developmental and disease contexts need to be better developed. Combining new tools in resolving dynamic transcriptomics paired with long read sequencing will help to discriminate these spatial-temporal patterns of different RNA isoforms in situ. In addition to transcriptomic and single-cell profiling in different contexts, it will be of great interest to understand the processing and turnover of lncRNAs in single cells, and to learn their suborganelle level localization patterns along with interacting proteins.
References:
-
Chen, Ling-Ling. "Towards higher-resolution and in vivo understanding of lncRNA biogenesis and function.” Nature methods vol. 19,10 (2022).
- Wang, Zhe et al. "LncRNA CRLM1 inhibits apoptosis and promotes metastasis through transcriptional regulation cooperated with hnRNPK in colorectal cancer.” Cell & bioscience vol. 12,1 120. 30 Jul. 2022.
- Huang, Zhao et al. "The role of long noncoding RNAs in hepatocellular carcinoma.” Molecular cancer vol. 19,1 77. 15 Apr. 2020.
- Yuan, Kefei et al. "Long noncoding RNA TLNC1 promotes the growth and metastasis of liver cancer via inhibition of p53 signaling." Molecular cancer vol. 21,1 105. 27 Apr. 2022.
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.


 Sample Submission Guidelines
Sample Submission Guidelines Overexpression of CRLM1 in colorectal liver metastases and associated with low survival rate
Overexpression of CRLM1 in colorectal liver metastases and associated with low survival rate CRLM1-OE converts CRC cell transcriptome into metastatic state
CRLM1-OE converts CRC cell transcriptome into metastatic state TLNC1 is upregulated in liver cancer tissues
TLNC1 is upregulated in liver cancer tissues TLNC1 interacts with TPR and represses the transcriptional activity of p53
TLNC1 interacts with TPR and represses the transcriptional activity of p53