We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy

The DNA Affinity Purification Sequencing (DAP-seq) technique is an innovative method aimed at exploring the interactions between transcription factors (TFs) and genomic DNA. By simulating the interaction between TFs and their DNA binding sites in vitro, this technology reveals key DNA sequences in gene regulatory networks. In contrast to traditional Chromatin Immunoprecipitation Sequencing (ChIP-seq), DAP-seq is not limited by antibody availability or species constraints, providing researchers with a more flexible research approach.
DAP-seq cleverly combines in vitro protein expression with high-throughput sequencing technology, eliminating the need for specific antibodies for each target TF. This approach allows for the comprehensive identification and analysis of transcription factor binding patterns throughout the entire genome in an environment unaffected by cellular chromatin structure. This is crucial for gaining deeper insights into complex transcriptional regulatory networks and their impact on organismal growth, development, and environmental adaptability.
Through DAP-seq, researchers can more precisely elucidate the molecular mechanisms of gene expression regulation and how environmental factors, such as DNA methylation, regulate these processes. The application of this technology not only advances our understanding of gene regulation mechanisms but also provides new perspectives and tools for future biomedical research and potential therapeutic strategies.
The cornerstone of DAP-seq is the replication of TFs naturally interacting with DNA within the cellular environment, albeit performed ex vivo. This groundbreaking methodology empowers researchers to directly observe and decipher TFs' genomic binding sites, unaffected by the complexities of the intracellular milieu. Central to DAP-seq's essence is the employment of an ex vivo expression system to engineer TFs fused with affinity tags. These tagged TFs are subsequently utilized to precisely isolate specific genomic binding sites through affinity purification techniques. Through meticulous examination of sequencing data, researchers can accurately pinpoint the genomic regions where TFs bind, thereby facilitating a comprehensive exploration of the intricate landscape of gene regulatory networks.
The DAP-seq methodology stands as a robust technique for discerning the binding sites of TFs throughout the entirety of the genome. This approach seamlessly integrates principles from molecular biology, biochemistry, and high-throughput sequencing technologies. Below is an elucidation of the meticulous steps constituting the DAP-seq workflow:
Genomic DNA Library Construction
In Vitro Protein Expression
Affinity Purification and Sequencing
Data Analysis
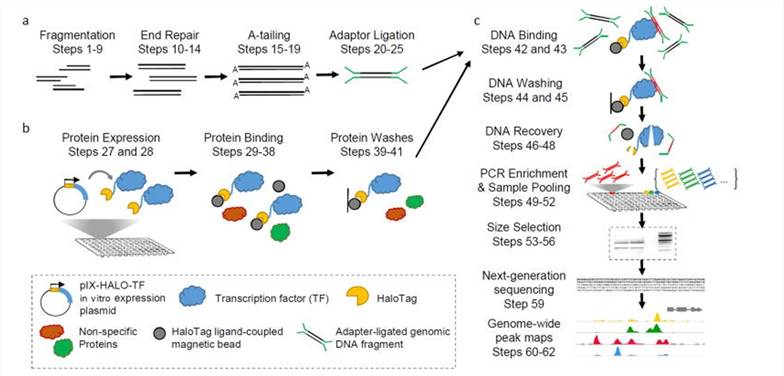 DAP-seq protocol overview(Bartlett et al., 2017)
DAP-seq protocol overview(Bartlett et al., 2017)
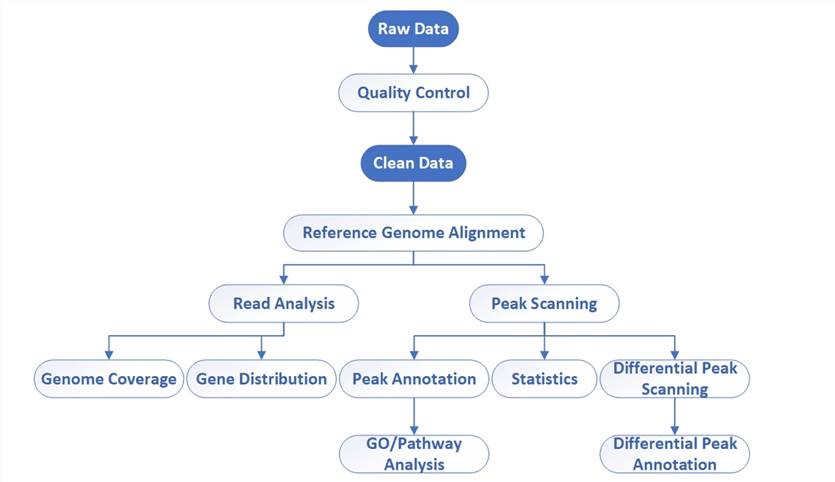 Bioinformatics workflow of DAP-Seq
Bioinformatics workflow of DAP-Seq
CD Genomics offers comprehensive and accurate DAP-seq technology services, with experienced expert teams performing quality control at each step of the process to ensure result accuracy.
Service you may intersted in
DAP-seq, an innovative method for probing the intricate dance between proteins and DNA, emerges as a formidable asset in the quest to map TF binding sites across the vast expanse of the genome. Its prowess lies in its precision, capable of pinpointing TF binding sites with remarkable accuracy across the entire genomic landscape, thereby offering a panoramic view of TF DNA binding specificity. For instance, researchers have harnessed the power of DAP-seq to chart a comprehensive map of TF binding sites throughout the Arabidopsis genome, shedding light on how DNA methylation influences TF binding dynamics. In comparison to traditional ChIP-seq approaches, DAP-seq dazzles with its heightened resolution, facilitating the precise delineation of TF binding regions sans the need for specific antibodies, thus alleviating concerns regarding antibody quality and nonspecific binding. Moreover, DAP-seq transcends the realm of TFs, delving into the binding characteristics of a myriad of DNA-binding proteins, thereby expanding its utility in unraveling molecular mysteries.
However, notwithstanding its advantages in identifying TF binding sites, DAP-seq grapples with certain limitations. The preparation of top-tier protein and DNA samples presents challenges, particularly when contending with proteins resistant to purification. Furthermore, despite its exceptional resolution, the specter of false positives and negatives looms, especially in the detection of low-abundance binding sites, resulting in variable success rates in identifying binding sites for different TF families. Additionally, the copious data output from DAP-seq necessitates sophisticated bioinformatics tools and expertise for robust data analysis and interpretation. Moreover, fine-tuning of DAP-seq protocols may be imperative for different proteins and biological samples to achieve optimal detection of binding sites.
In summation, while DAP-seq stands as a potent instrument for unraveling the regulatory ballet orchestrated by TFs in governing gene expression, its effective deployment mandates meticulous sample preparation, rigorous data analysis, and potential refinements in technique.
References
Terms & Conditions Privacy Policy Copyright © CD Genomics. All rights reserved.