Epigenetics, the study of heritable changes in gene expression that do not involve alterations to the DNA sequence, encompasses phenomena that result in inheritable phenotypic modifications. Among the various mechanisms of epigenetic regulation, DNA methylation is the most extensively investigated and understood. DNA methylation, a common mode of gene expression regulation, plays a crucial role in numerous biological processes. Aberrant DNA methylation, including hypermethylation and hypomethylation, can significantly impact biological processes such as cancer, neurological disorders, aging, and development.
To elucidate the DNA methylation landscape of the human genome, Illumina has developed microarray products, including the Infinium™ MethylationEPIC BeadChips (850K), Infinium Human Methylation 935K and Infinium Methylation Screening Array 270K designed to assess methylation levels across genomic loci. However, there is a scarcity of high-performance, cost-effective microarrays for model organisms. In some studies, researchers have even utilized human DNA methylation arrays to investigate murine methylation patterns .
In response to this need, Illumina has introduced the Infinium™ Mouse Methylation BeadChip, specifically designed for the quantitative analysis of DNA methylation in the model organism, Mus musculus. This microarray can simultaneously analyze 12 samples, with each sample covering up to 285,000 CpG sites. The array provides comprehensive genomic coverage, including CpG islands, transcription start sites (TSS), enhancers, imprinting control regions, gene bodies, repetitive elements, lamin-associated domains, CTCF binding sites, and hypermethylated regions in cancer. Furthermore, this array is suitable for methylation analysis across various mouse strains, making it the preferred choice for murine methylation research (Figure 1)
 Figure 1: Illumina Infinium™ Mouse Methylation BeadChip
Figure 1: Illumina Infinium™ Mouse Methylation BeadChip
Principle of the Infinium™ Mouse Methylation BeadChip
The Infinium™ Mouse Methylation BeadChip utilizes the same design principles as the Infinium™ Human Methylation450K and 850K BeadChips, incorporating both Infinium I and Infinium II probe types.
Infinium I Probes: Each methylation site is interrogated by two distinct probes. The M-probe, which ends in a guanine (G), is designed to detect the methylated cytosine (C) at the target site. The U-probe, ending in an adenine (A), is intended to detect the unmethylated thymine (T) at the same locus (Figure 2). According to the principle of single-base extension, a fluorescently labeled dideoxynucleotide triphosphate (ddNTP) is incorporated only when the final base of the probe matches the template DNA. The methylation value is then calculated based on the fluorescent signals from both the M and U probes.
Infinium II Probes: These probes utilize a single bead type to detect methylation status. The incorporation of either an adenine (A) or a guanine (G) base at the extension site determines the methylation state. If an adenine is incorporated, the site is unmethylated; if a guanine is incorporated, the site is methylated. The methylation level at the target site is then quantified by analyzing the signal intensities corresponding to the different incorporated bases (Figure 2).
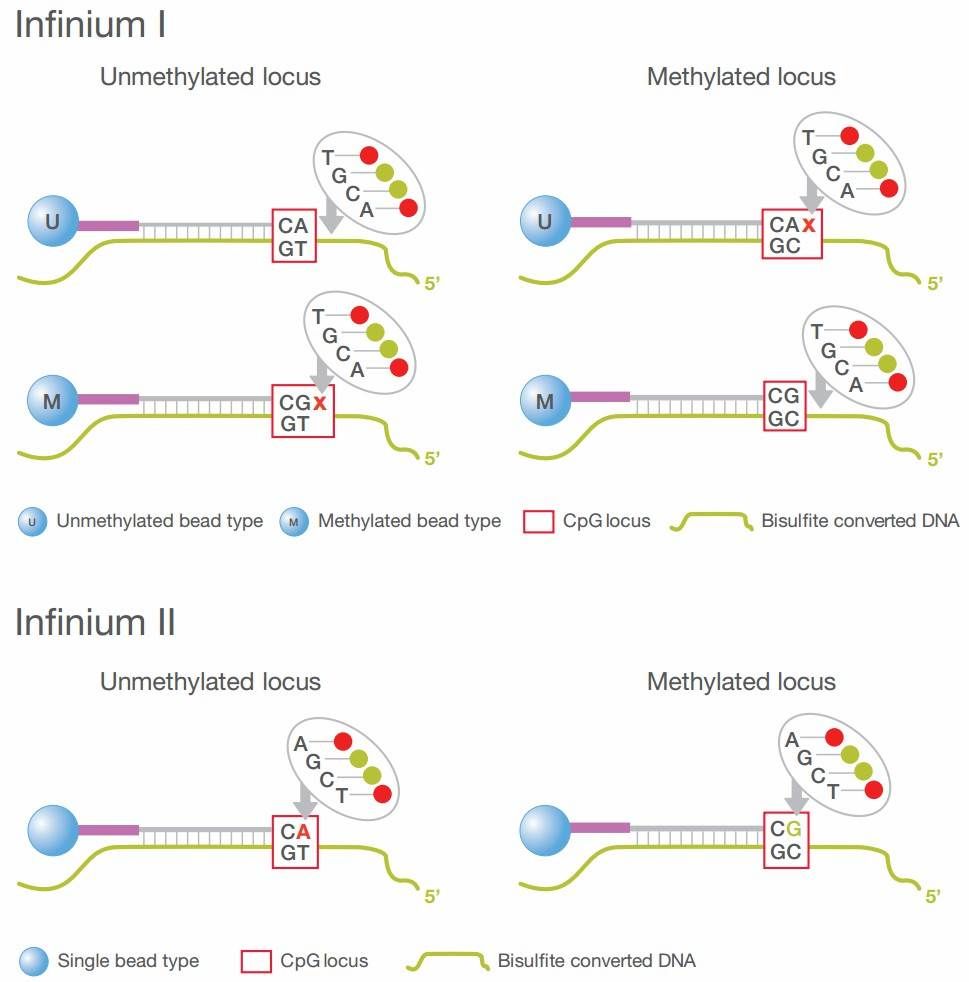 Figure 2: Design Principles of Infinium I and Infinium II Probes
Figure 2: Design Principles of Infinium I and Infinium II Probes
Service Offerings for Infinium™ Mouse Methylation BeadChip by CD Genomics
CD Genomics provides comprehensive services for the Infinium™ Mouse Methylation BeadChip, catering to researchers engaged in DNA methylation studies. CD Genomics is equipped with multiple platforms, including iScan, Thermo Fisher, MassARRAY, and KASP, offering a wide range of chip types such as GSA, GSA+MD, ASA, ASA+MD, ASA-CHIA, OmniChip, and the 850K Methylation Chip. Researchers are encouraged to consult with CD Genomics for expert support in their DNA methylation research endeavors.
Technical Features of the Infinium™ Mouse Methylation BeadChip
Extensive Genome-Wide Coverage: The Infinium™ Mouse Methylation BeadChip interrogates over 285,000 methylation sites, encompassing CpG islands, transcription start sites (TSS), enhancers, imprinting control regions, gene bodies, repetitive sequences, lamin-associated domains, CTCF binding sites, and regions of hypermethylation in cancer (Table 1, Figure 3).
Table 1. Distribution of 285,000 Methylation Sites in the Genome
| Feature | Description of Coverage |
| TSS within 500 bp of protein-coding transcripts | Covers over 28,000 protein-coding transcripts. |
| miRNA promoter TSS | Covers all annotated miRNA from Ensembl v75; includes over 1,900 miRNAs in total. |
| Noncoding RNA TSS | Covers lncRNA sites as annotated by Ensembl v75. |
| Enhancers | Sites defined by VISTA and mouse ENCODE; includes over 60,000 enhancers in total. |
| Imprinted loci | Sites selected by whole-genome bisulfite sequencing (WGBS) where intermediate methylation is observed across various cell types. |
| CpG islands | Selected from University of California, Santa Cruz (UCSC) mouse CpG island collection where TSS does not overlap with CpG island; approximately 16,000 CpG islands in total. |
| Hypermethylated regions in cancer | Selected from WGBS data of mouse tumors and normal tissue; includes over 5,700 CpG sites in total. |
| Non-CpG cytosine probes (CpH sites) | CpH sites evenly distributed by sequence context to CAG, CTG, CCG, CAH, CTH, and CCH sites; approximately 4,000 sites in total. |
| CpGs from consensus repetitive elements | Selected for conserved sequences for Line1, B1, and IAPEYI elements. |
| Gene body sites | Random sites more than 2 kb from protein TSS. |
| CTCF binding sites | Selected from sequences that overlap CTCF ChIP-Seq peaks from Mouse ENCODE. |
| Human MethylationEPIC liftover | Selected from the most highly conserved Human MethylationEPIC probes mapped onto the mouse genome. |
| Random CpG probes | Sites randomly selected to reduce bias in the assessment of global genomic DNA methylation. |
| Strain-specific SNP probes | One probe from each of the 20 chromosomes for the 18 mouse strains covered by the mouse genome project. |
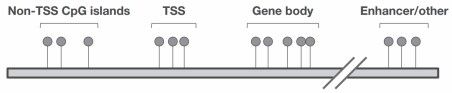 Figure 3. Coverage Regions of the Infinium™ Mouse Methylation BeadChip
Figure 3. Coverage Regions of the Infinium™ Mouse Methylation BeadChip
Simplified Workflow: The workflow of the Infinium™ Mouse Methylation BeadChip is streamlined, eliminating the need for PCR and requiring as little as 250 ng of starting DNA, making it suitable for precious samples with limited DNA availability. The chip kit includes all necessary reagents for methylation analysis, excluding bisulfite conversion reagents, thus facilitating straightforward experimental procedures.
Robust Chip Design: The Infinium™ Mouse Methylation BeadChip employs both Infinium I and II analytical chemistries (Figure 2). This dual approach ensures data stability and reliability while maximizing detection coverage. Infinium II utilizes degenerate oligonucleotides as probes on individual beads, maintaining accuracy even in the presence of up to three methylated CpG sites within the sequence.
Reliable Chip Data: The stability and reliability of chip data are evidenced by technical replicates. For the Infinium™ Mouse Methylation BeadChip, the technical replicate correlation (R^2) exceeds 0.98 (Figure 4), indicating high data consistency. Additionally, the comparison of biological replicates reveals a delta-β value of 0.2, with a false positive rate of less than 1%.
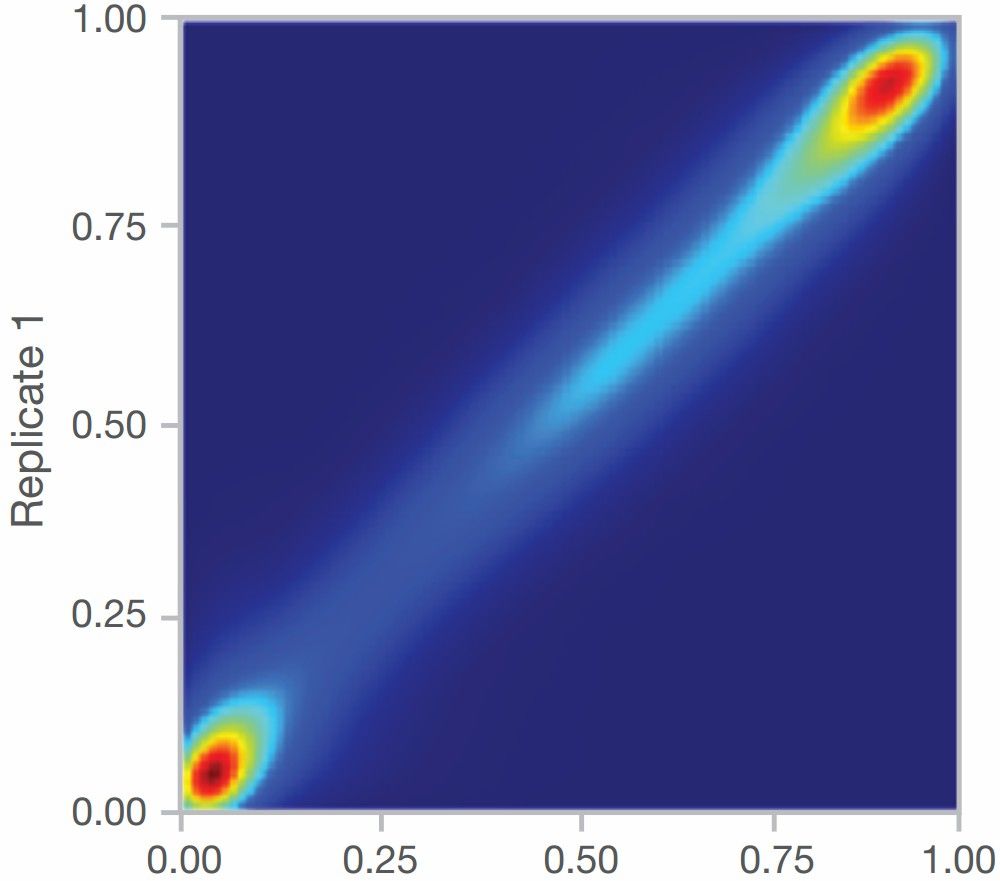 Figure 4. Technical Replicate Correlation of the Infinium™ Mouse Methylation BeadChip (β values)
Figure 4. Technical Replicate Correlation of the Infinium™ Mouse Methylation BeadChip (β values)
Stringent Internal Quality Control: The Infinium™ Mouse Methylation BeadChip includes negative control sites to account for reduced sequence complexity post-bisulfite conversion. The GenomeStudio™ Methylation Module software features an integrated Controls Dashboard for convenient monitoring of controls.
Integrated and Compatible Analysis Software: GenomeStudio, developed by Illumina, supports data analysis for the Infinium™ Mouse Methylation BeadChip, enabling researchers to conduct differential methylation analyses with ease. GenomeStudio also provides advanced visualization tools, allowing researchers to view extensive data sets in single images, such as heatmaps and scatter plots (Figure 5). Methylation results from iScan are compatible with multiple third-party software and various R packages for data normalization and differential analysis.
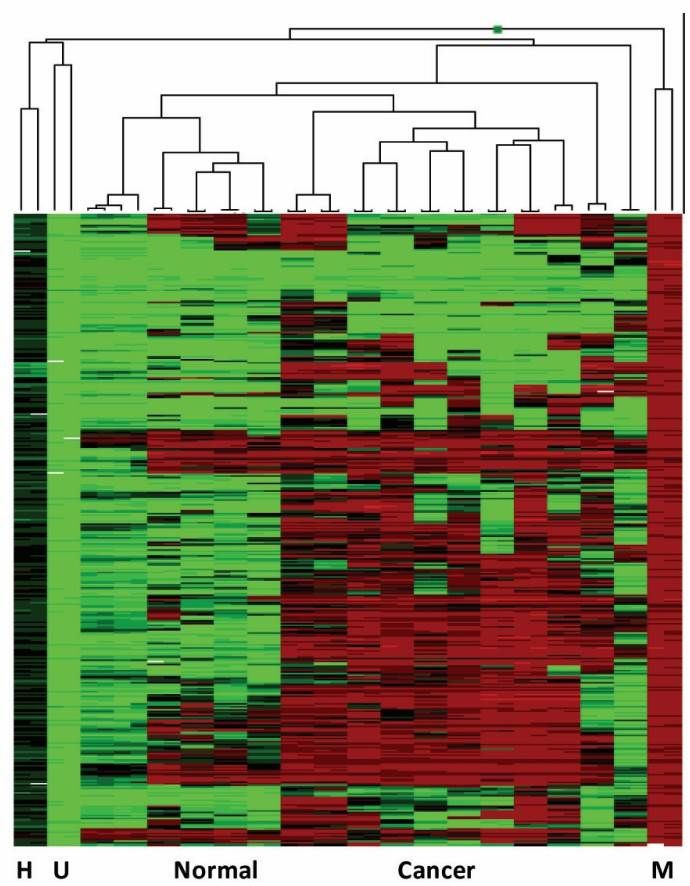 Figure 5. Heatmap Generated by GenomeStudio
Figure 5. Heatmap Generated by GenomeStudio
Wide Range of Applications: With over 200,000 modified mouse strains developed, primarily for modeling human diseases, the Infinium™ Mouse Methylation BeadChip has extensive applications. These include epigenome-wide association studies (EWAS), tumor xenograft models, preclinical research, toxicological studies, developmental biology, and any research involving DNA methylation-based studies. The chip is suitable for validation work across these diverse areas.
The Infinium™ Mouse Methylation BeadChip thus provides a comprehensive, robust, and reliable platform for the quantitative analysis of DNA methylation in murine models, supporting a wide range of research applications with high data integrity and simplified workflows.
Data Analysis
The bioinformatics analysis conducted by CD Genomics as part of the Mouse DNA Methylation Microarray Service is pivotal in transforming raw data into valuable biological insights. Leveraging advanced computational tools and robust algorithms, this analysis provides a comprehensive understanding of DNA methylation dynamics and its potential implications.
Sample Preparation
Total Sample Quantity: Each sample should have a total amount of no less than 250 ng.
Sample Concentration: The minimum concentration should be no less than 50 ng/μL.
Sample Purity: The OD 260/280 ratio should be between 1.7 and 1.9.
Sample Quality: The genomic DNA should be intact, free from degradation and RNA contamination, and devoid of other genomic DNA contaminants.
Sample Transportation: DNA should be transported at low temperatures (-20°C).
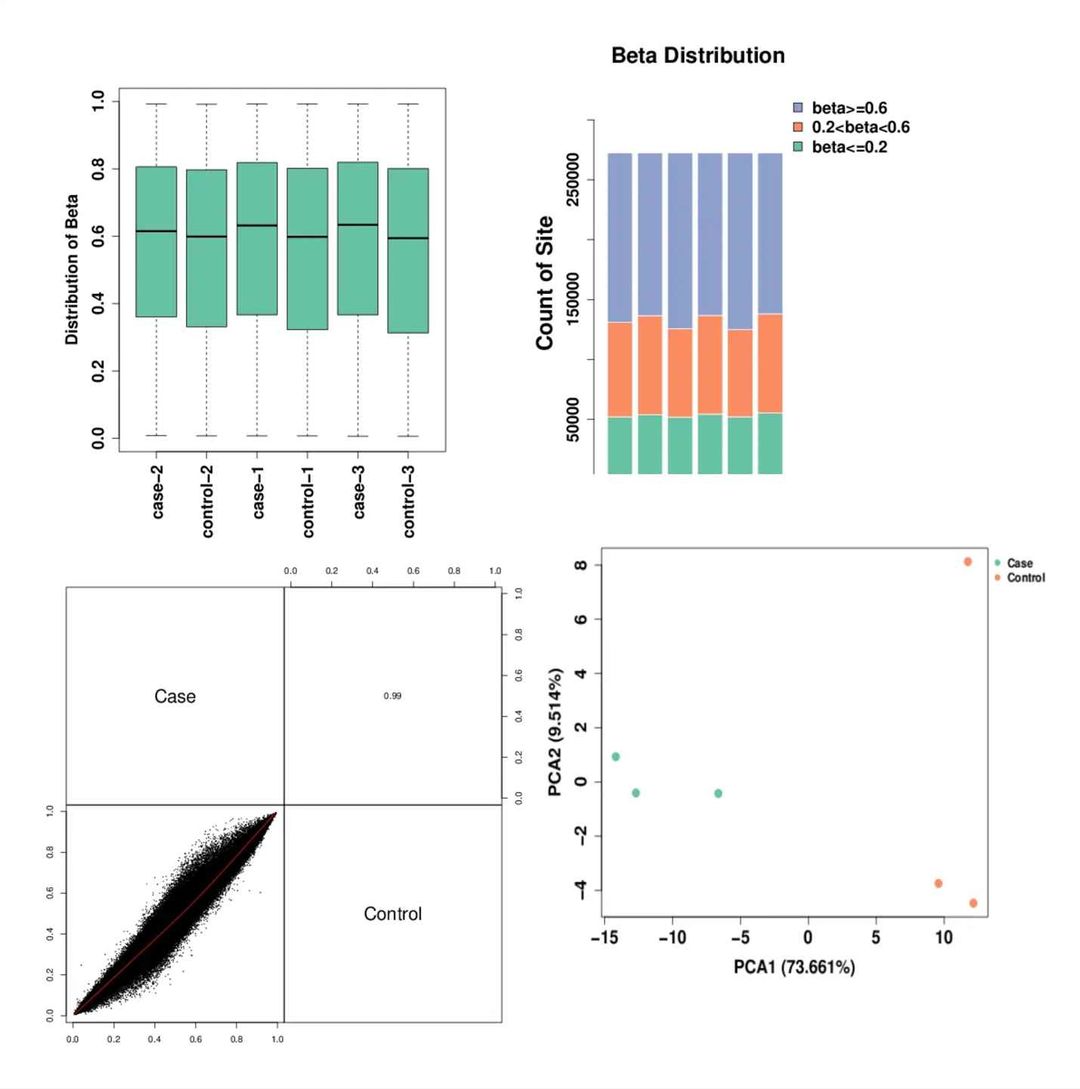
Methylation Levels and Group Relationship Diagram
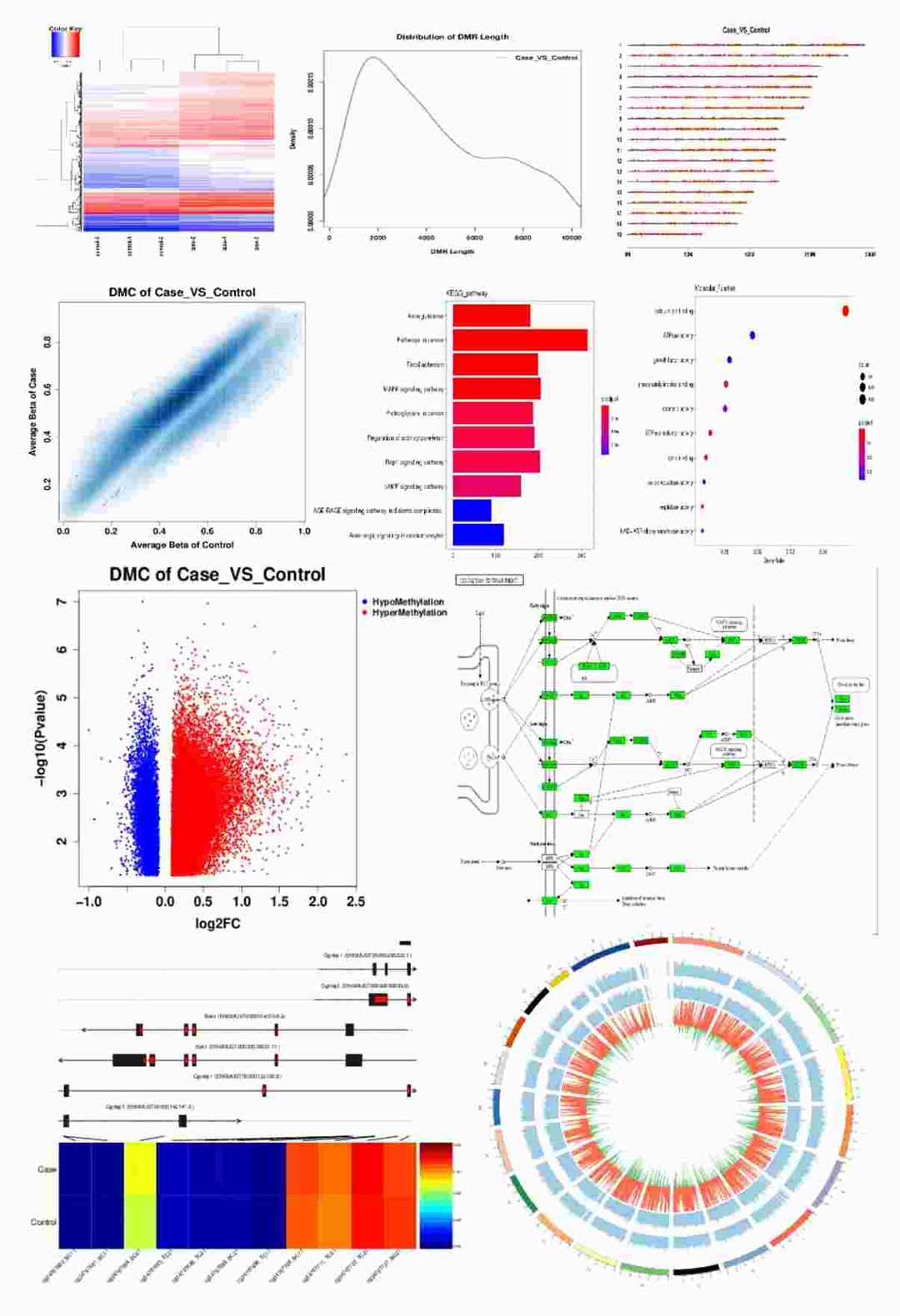
Differential Methylation Screening and Downstream Enrichment Analysis Diagram
References
- Wong, N. C., Ng, J., Hall, N. E., Lunke, S. , Salmanidis, M. , & Brumatti, G. , et al. (2013). Exploring the utility of human dna methylation arrays for profiling mouse genomic dna. Genomics, 102(1), 38-46.
- Gujar H, Liang JW, Wong NC, Mozhui K (2018) Profiling DNA methylation differences between inbred mouse strains on the Illumina Human Infinium MethylationEPIC microarray. PLoS ONE 13(3): e0193496.
- https://www.illumina.com
