Human Illumina Methylation Assay (935K) Service
Illumina DNA methylation plays an important role in gene expression regulation, and changes in the DNA methylation status of cellular DNA are associated with aging, development, and pathological conditions. Over the past decade, Illumina has provided researchers with powerful microarray-based tools driven by BeadArray™ technology to quantitatively analyze DNA methylation across the entire genome. So far, the Infinium Human Methylation 450 (450K) and Methylation EPIC v1.0 BeadChips (850K) have been widely used in the research community to facilitate data collection for epigenome-wide association studies (EWAS). These methylation arrays have enabled the discovery and application of DNA methylation-based biomarkers in cancer research, genetic diseases, aging, and molecular epidemiology. Building upon this foundation, Illumina has introduced the next generation of upgraded methylation array, the Infinium MethylationEPIC v2.0 BeadChip (935K), which is based on the same reliable Infinium chemistry but incorporates updates and optimizations of many methylation sites, aiming to enable more biological discoveries in the era of epigenetics research.
The Infinium MethylationEPIC v2.0
BeadChip Kit (935K) is a whole-genome DNA methylation screening tool that targets over 935,000 CpG sites in the most biologically significant regions of the human methylome. It significantly improves backward compatibility while retaining the ability to quantitatively analyze genome-wide CpG at single-nucleotide resolution and providing highly accurate and precise methylation measurements unaffected by sequencing depth. The kit enables streamlined and user-friendly Infinium methylation detection and analysis of various DNA sample types, including DNA isolated from FFPE (formalin-fixed, paraffin-embedded) samples. Compared to other methods, MethylationEPIC v2.0 offers scalability and lower per-sample costs, making it suitable for investigating disease mechanisms and biomarker screening.
Characteristics of the 935K Chip
Whole-genome coverage and upgraded CpG site detection:
The Illumina EPICv2.0-935K chip can detect the methylation status of approximately 935,000 CpG sites in the human genome. It improves upon the 850K chip by removing underperforming probes and introducing the following enhancements:
(1) Addition of 186,000 CpG sites targeting enhancer and super-enhancer regions.
(2) Increased coverage of CTCF binding sites, CNV detection regions, CpG islands with insufficient coverage in EPIC v1.0, and common cancer driver mutations.
(3) Inclusion of chromatin open regions associated with tumors identified through ATAC-Seq and ChIP-Seq experiments.
(4) Updated genome version to hg38 and GenCode database version v41. This chip is more suitable for epigenome-wide association studies (EWAS) in the analysis of the entire epigenome.
Compatibility with various sample types, including FFPE:
High-quality data can be obtained from FFPE tissue samples. FFPE compatibility is crucial for cancer research and supports studies based on large tumor biobanks. The 935K chip is compatible with the following sample types: FFPE, fresh/frozen tissue, whole blood, buccal cells, cfdna (cell-free DNA), saliva and other sample types.
Highly accurate and precise detection data:
The 935K methylation chip can detect differences as small as 0.2 in beta values with a false positive rate of less than 1%, achieving high analytical sensitivity and excellent data reproducibility. As shown in the following figures: (A) Technical replicates of the 935K chip show a correlation R2>0.99. (B) The correlation between the probes shared by the 935K and 850K chips is R2>0.99. (C) The correlation between the data from the 935K chip and methylation sequencing (at 100× sequencing depth) is R2>96%.
Service Advantages:
Mature service platform: A stable and systematic chip experiment solution that provides customers with total DNA extraction, sample quality control, RNA labeling, hybridization, elution, image scanning, and data analysis.
Diversified service offerings: Provides both chip-based and sequencing-based high-throughput detection technologies, as well as low-throughput detection technology RT-PCR.
Professional laboratory personnel: Professional laboratory personnel are available for DNA extraction and chip hybridization, ensuring the smooth completion of chip experiments with their rich experience in chip operations.
Professional analysis team: An experienced analysis team that can quickly complete basic analysis and provide personalized analysis according to customer needs, offering various high-quality charts and figures.
CD Genomics has accumulated extensive project experience in methylation chip detection, handling various types of human samples, including blood, tissue, and FFPE samples.
935K Chip Advantages
The 935K chip is an upgraded version of the 850K chip and offers the following advantages:
(1) Comprehensive genome coverage: Detects over 935,000 CpG sites, covering CpG islands, promoters, coding regions, and enhancers comprehensively.
(2) Comprehensive optimization of probe sites: Removes poorly performing probes from EPICv1.0-850K and adds 186,000 CpG-targeting enhancers and super-enhancers, more CTCF binding sites, CNV detection regions, CpG islands not well covered by EPIC v1.0, and common cancer-driving mutations.
(3) Compatibility with various sample types, including FFPE.
(4) High-quality data: Adopts the Infinium technology from the 450K chip and utilizes Infinium I and II probe designs to maximize the detection range.
(5) High resolution: Single-base resolution that directly detects accurate methylated sites.
(6) High reproducibility: Strong correlation between technical replicates and R2>0.99 correlation with the same probes as the 850K chip.
The 935K Chip Application
(1) Tumors: lung cancer, esophageal cancer, glioma, leukemia, etc.
(2) Complex diseases: diabetes, hypertension, immune disorders, depression, etc.
(3) Development: embryonic development, neural differentiation, organ differentiation, etc.
(4) Early screening: tumor biomarkers, recurrence markers, prognostic markers.
(5) Aging.
Sample Requirements:
Sample types: Tissue, cells, genomic DNA.
Sample purity: OD 260/280 value should be between 1.7 and 2.0; RNA should be thoroughly removed; no contamination from other individuals or species' DNA.
Sample concentration: The concentration should not be lower than 50ng/μl.
Sample amount: Each sample should have a total amount of no less than 500ng. Sample solvent: Dissolve in TE.
Sample transportation: DNA should be transported at a low temperature (-20°C); during transportation, seal the tube opening with parafilm to prevent contamination.
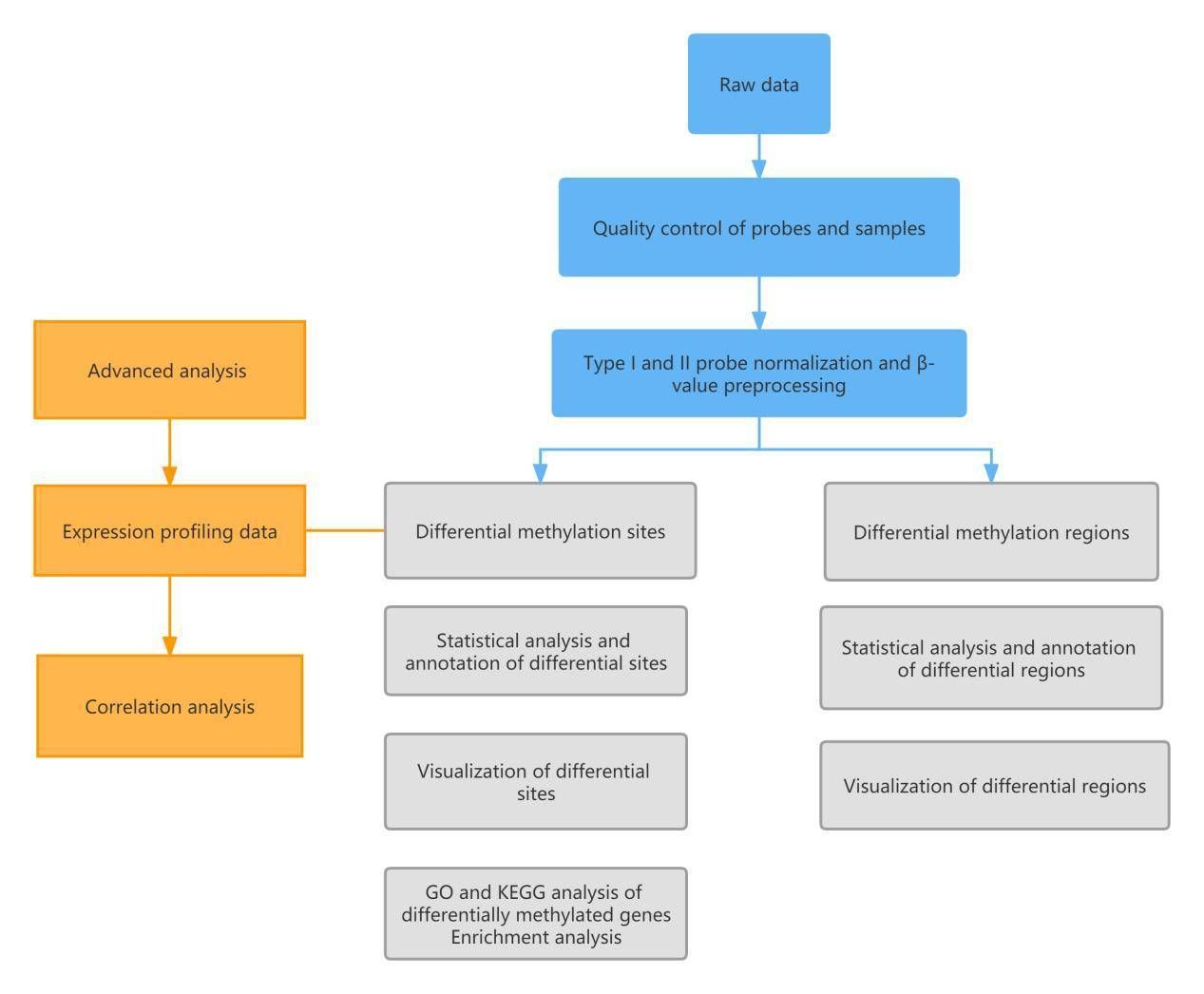
Analysis workflow