DNA methylation plays a crucial role in gene expression regulation and is involved in various important biological processes. Moreover, aberrant DNA methylation and its impact on gene expression can lead to the development of complex human diseases, such as cancer, neurological disorders, and diabetes. In order to study the significant role of methylation in the human body and accelerate the clinical progress of methylation research, Illumina introduced the high-throughput 450k DNA methylation microarray, which has achieved tremendous success. Building upon this foundation, Illumina has launched the next-generation upgraded version, the Infinium MethylationEPIC BeadChip (referred to as the 850k chip), providing researchers with a more cost-effective and reliable platform for methylation studies.
The Illumina Infinium MethylationEPIC BeadChip (850K chip) provides researchers with a reliable and cost-effective platform for methylation analysis. It has improved the protocol for detecting FFPE samples to obtain more reliable and stable results. Widely used in stem cell research, tumor studies, and other complex disease research, it is currently the preferred whole-genome DNA methylation microarray for epigenomic-wide association studies.
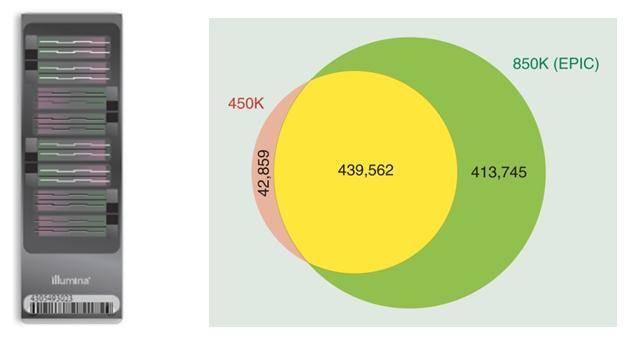
The 850K chip can detect the methylation status of approximately 853,307 CpG sites in the human genome, which includes 91% of the sites covered by the original 450K chip and an additional 413,745 sites. The 850K chip not only maintains comprehensive coverage of CpG islands and gene promoter regions but also enhances the coverage of enhancer regions (adding 333,265 probes covering enhancers from the ENCODE and FANTOM5 projects) as well as probes covering gene coding regions.
CD Genomics offers human DNA methylation Assay services utilizing off-the-shelf platforms such as Illumina, Agilent, and more. We will aid you in epigenome wide association studies by combining extensive, expert-selected coverage and high-throughput compatible with big sample size using the industry's highly trusted, validated DNA analysis systems.
Infinium MethylationEPIC BeadChip (850K chip)
The Infinium MethylationEPIC BeadChip, also known as the 850K chip, can detect the methylation status of approximately 853,307 CpG sites in the human genome. It includes 91% of the sites covered by the original 450K chip and adds an additional 413,745 sites. The 850K chip not only maintains comprehensive coverage of CpG islands and gene promoter regions but also significantly enhances coverage of enhancer regions, incorporating the latest research advancements from ENCODE and FANTOM5 projects, as well as probe coverage in gene coding regions. It is widely applicable in stem cell research, tumor studies, and other investigations of complex diseases. The 850K chip is particularly suitable for epigenomic-wide association studies (EWAS) and comprehensive analysis of the epigenome-genome relationship.
850K Chip Coverage:
Covers over 90% of the sites from the Infinium Methylation 450 BeadChip.
Includes CpG sites outside of CpG islands.
Covers non-CpG methylation sites (CHH sites) in human stem cells.
Includes differentially methylated sites between normal and tumor tissues (various types of cancer).
Covers FANTOM5 enhancers.
Covers open chromatin and enhancers from the ENCODE project.
Includes ultra-sensitive sites for DNAse hypersensitivity.
Covers miRNA promoter regions.
Workflow of Infinium methylation Assay
Advantages of The 850K Chip:
Comprehensive genomic coverage: Detects over 850,000 CpG sites, providing comprehensive coverage of CpG islands, promoters, coding regions, and enhancers.
High-quality data: Utilizes the Infinium technology from the 450K chip and incorporates both Infinium I and II probe designs, maximizing the detection range.
High resolution: Single-base resolution allows for precise identification of methylated sites.
High reproducibility: Strong correlation between technical replicates and with the 450K chip probes (R2 > 0.98).
Low starting template requirement: Only 250 ng of DNA is needed, resulting in significant sample savings.
Suitable for FFPE samples.
Our Human DNA Methylation Assay Service Advantages
Mature service platform: We provide a stable and comprehensive solution for chip experiments, offering services such as total DNA extraction, sample quality control, RNA labeling, hybridization, washing, image scanning, and data analysis.
Diverse service offerings: We provide both chip-based and sequencing-based high-throughput technologies, as well as low-throughput RT-PCR technology.
Professional laboratory staff: Our team of experienced laboratory personnel ensures smooth execution of chip experiments, from DNA extraction to chip hybridization.
Expert analysis team: Our analysis team has extensive practical experience and can quickly perform basic analysis. We also offer personalized analysis based on customer requirements, providing various high-quality charts and figures.
Sample requirements:
Sample types: Tissues, cells, genomic DNA.
Sample purity: OD 260/280 value should be between 1.7 and 2.0. RNA should be thoroughly removed, and there should be no contamination of DNA from other individuals or species. Sample concentration: Concentration should not be lower than 50 ng/μl.
Sample quantity: Each sample should have a total quantity of at least 500 ng. Sample solvent: Dissolve the sample in TE buffer. Sample transportation: DNA should be transported at a low temperature (-20°C). During transportation, please seal the tube with parafilm to prevent contamination.
The Journal of Clinical Oncology reported on the predictive methylation site markers for postoperative recurrence in early-stage liver cancer.
The study used the 450K chip to analyze 66 patient samples and filtered out 2550 differential sites. By comparing the differential CpG sites obtained from two algorithms, a total of 46 CpG sites were selected. Subsequently, the researchers conducted testing on an internal sample group and two external sample groups. To establish a clinically applicable model for predicting individual recurrence, the researchers used nomograms considering covariates and successfully built a predictive model with three calibration points yielding ideal results. The researchers developed a nomogram to predict the postoperative recurrence risk in early-stage liver cancer patients.
Reference
- Qiu J, Peng B, Tang Y, et al. CpG methylation signature predicts recurrence in early-stage hepatocellular carcinoma: results from a multicenter study[J]. J Clin Oncol, 2017, 35(7): 734-42.
