Gene expression can be regulated by changing the topological structure of chromatin or chromatin modification. The dynamic remodeling of chromatin is closely related to embryonic development, cell aging, tumorigenesis, immunity, and cell fate determination. In addition, the dynamic structure of chromatin is also related to human health. Chromatin Accessibility, defined by the openness of chromatin, has an important impact on the expression of genes, DNA replication, repair and other life activities of cells, and is always strictly regulated by cells, which reflects the transcriptional activity of chromatin, is an important direction for studying gene expression regulation, and plays an important role in the mapping of epigenetic map, cell differentiation and development, and the occurrence and development of various diseases.
Single cell sequencing and ATAC-seq applied to map human genome single-cell chromatin accessibility
Cells in various organs and tissues of human body are highly specialized, and their specialized functions are regulated by CRE. To profile the activity of gene regulatory elements in diverse cell types and tissues in the human body, researchers applied single-cell chromatin accessibility assays to 30 adult human tissue types from multiple donors. The author analyzed about 600000 cells from 30 human tissues using single-cell sequencing technology and the potential CRE in cells was identified by using ATAC-seq. Based on the integrated analysis of the obtained results and previous data, the largest human single-cell chromatin accessibility map has been drawn, and nearly 1.2 million cis-acting elements have been annotated in 222 cell subtypes, providing valuable reference resources for the analysis of gene transcription regulation mechanism and the pathogenesis of human complex diseases. This study provides valuable reference resources for analyzing the mechanism of gene transcription regulation and the pathogenesis of human complex diseases.
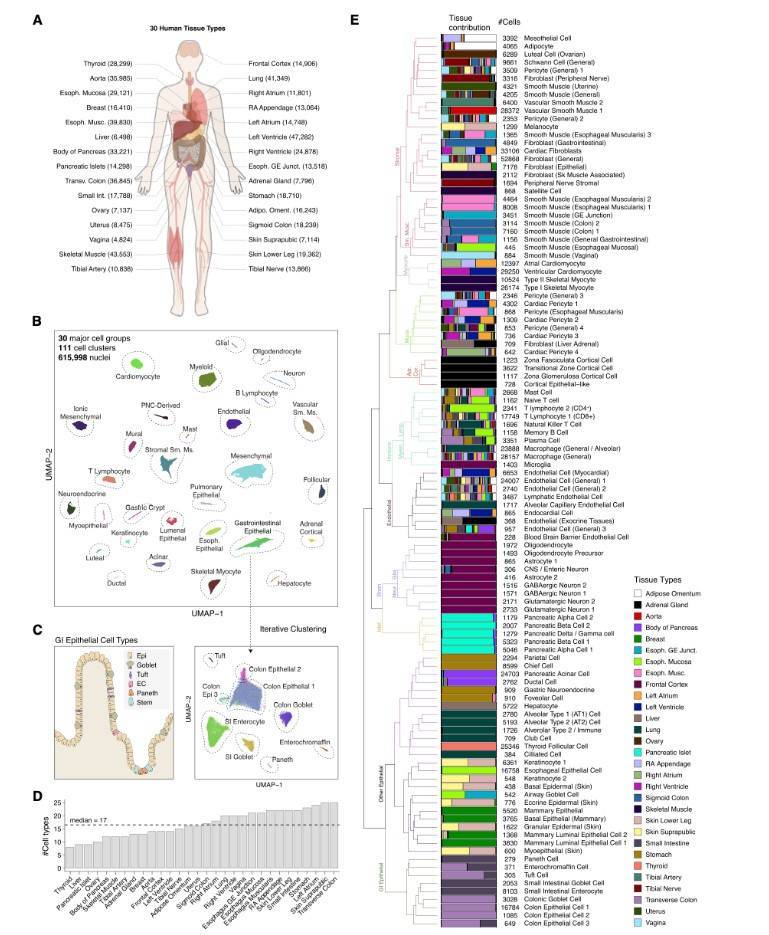 Analysis of single cell chromatin accessibility in 30 adult primary tissues used for sci-A TAC-seq
Analysis of single cell chromatin accessibility in 30 adult primary tissues used for sci-A TAC-seq
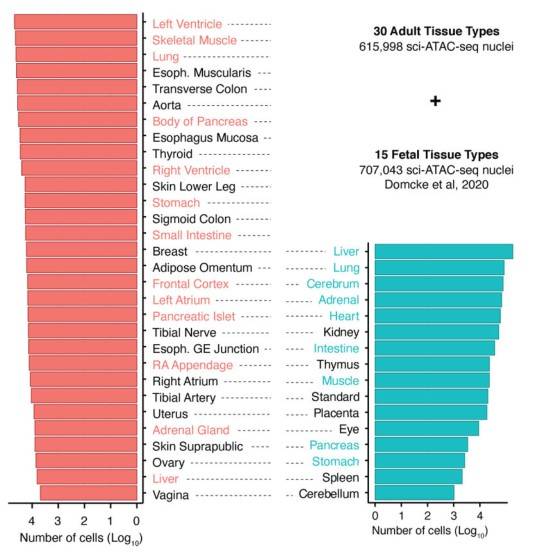 Number of sci-A TAC-seq cells per tissue type for 30 adult and 15 human fetal tissue types that were integrated.
Number of sci-A TAC-seq cells per tissue type for 30 adult and 15 human fetal tissue types that were integrated.
Analysis of single cell atlas of dynamic chromatin of immunocytes in renal cell carcinoma by multi-group method
Renal cell carcinoma (ccRCC) is a tumor that responds to immunotherapy. The development of immune checkpoint inhibitors has changed the management of advanced RCC. Nonetheless, the majority of patients either have primary resistance to therapies or develop resistance after an initial response. RCC displays unique characteristics compared with other immune-responsive solid tumors, including a modest mutation burden and association of increased infiltration of CD8+ T cells with worse prognosis. Consequently, in order to develop new and improved immune-based RCC treatment methods, it is essential to understand the development and functional status of patients' immune cells. A recent study employs single-cell assay technology for transposase-accessible chromatin using sequencing to study T cells' chromatin map in normal and tumor tissues of patients with renal cell carcinoma. Furthermore, all 8 patients were performed with scATAC-seq, of which 4 patients were performed with matched scATAC-seq and scRNA-seq, and matched transcriptome sequencing and TCR sequencing were performed in the part of scRNA-seq. The data show that intratumoral CD8+ T cells demonstrates a continuum of dysfunctional states and an extensive remodeling of the accessibility of regulatory elements through surveying the chromatin landscapes of T cells of malignant and nonmalignant tissues from patients with ccRCC. This study provides a valuable resource for dissecting the epigenetic and transcriptional heterogeneity of T cells in ccRCC and these comprehensive T cell single cell maps help to understand the T cell biology of cancer patients, and can guide treatment strategies to overcome drug tolerance caused by immune cell heterogeneity.
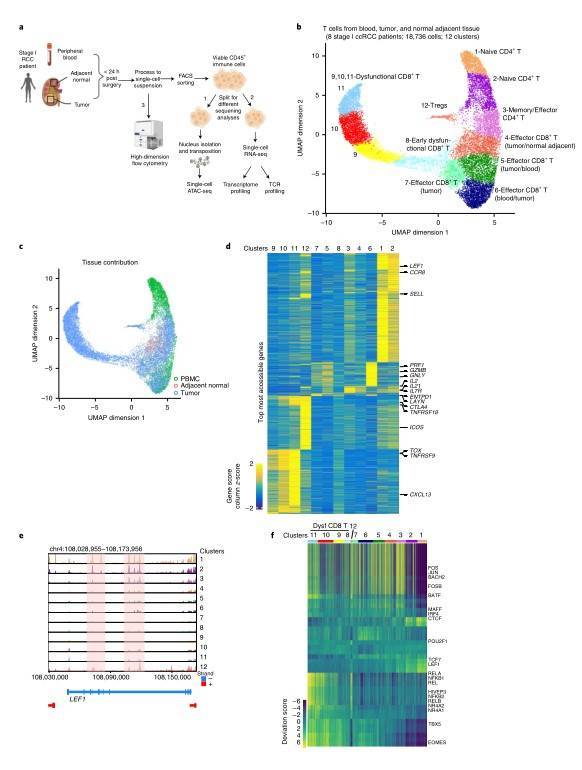 Single-cell chromatin accessibility of T cells in ccRCC
Single-cell chromatin accessibility of T cells in ccRCC
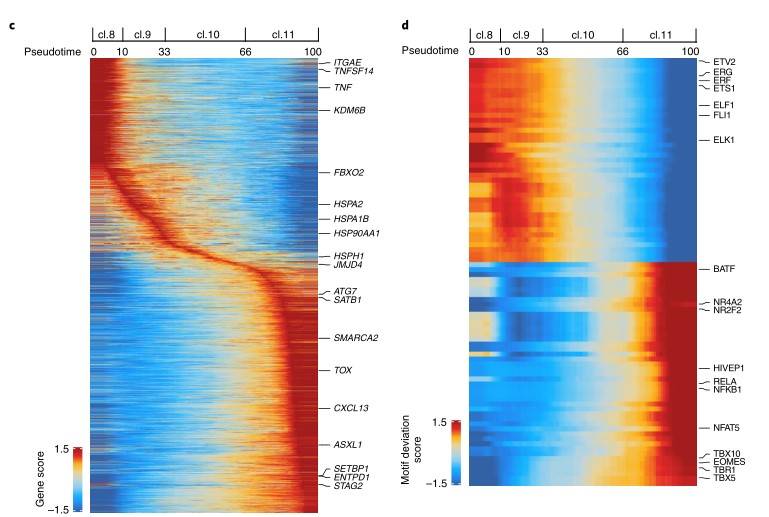 Workflow for integrating scATAC-seq and scRNA-seq data from the same samples
Workflow for integrating scATAC-seq and scRNA-seq data from the same samples
With a global understanding of chromatin accessibility at the genome-wide level, we can decipher the effective regulatory elements in gene transcription regulation and provide new ideas for further understanding the pathogenesis of diseases.
References:
- Zhang, Kai et al. "A single-cell atlas of chromatin accessibility in the human genome." Cell vol. 184,24 (2021): 5985-6001.e19.
- Tang, Fanying et al. "Chromatin profiles classify castration-resistant prostate cancers suggesting therapeutic targets." Science (New York, N.Y.) vol. 376,6596 (2022): eabe1505.
- Kourtis, Nikos et al. "A single-cell map of dynamic chromatin landscapes of immune cells in renal cell carcinoma." Nature cancer vol. 3,7 (2022): 885-898.
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.


 Sample Submission Guidelines
Sample Submission Guidelines
 Analysis of single cell chromatin accessibility in 30 adult primary tissues used for sci-A TAC-seq
Analysis of single cell chromatin accessibility in 30 adult primary tissues used for sci-A TAC-seq  Number of sci-A TAC-seq cells per tissue type for 30 adult and 15 human fetal tissue types that were integrated.
Number of sci-A TAC-seq cells per tissue type for 30 adult and 15 human fetal tissue types that were integrated. Single-cell chromatin accessibility of T cells in ccRCC
Single-cell chromatin accessibility of T cells in ccRCC  Workflow for integrating scATAC-seq and scRNA-seq data from the same samples
Workflow for integrating scATAC-seq and scRNA-seq data from the same samples