Introduction
The discovery of tumor neoantigens holds immense promise for the development of effective individualized immunotherapies, including cancer vaccines. Traditionally, neoantigen discovery has primarily focused on single nucleotide variants (SNVs) using short-read sequencing technologies. However, these approaches often overlook critical alterations in transcript structures that arise from alternative splicing events, which are frequently observed in cancer. To address this limitation and unlock deeper insights into tumor heterogeneity, a groundbreaking study titled "An Isoform-Resolution Transcriptomic Atlas of Colorectal Cancer: Unveiling Heterogeneities and Novel Neoepitopes through Integrated Long-Read Single-Cell Sequencing" provides valuable insights into the complex landscape of tumor heterogeneity and the discovery of novel neoepitopes.
Integration of Short- and Long-Read Sequencing
The study employed an innovative approach that combined short-read single-cell RNA sequencing (scRNA-seq) with long-read scRNA-seq to create an isoform-resolution transcriptomic atlas of colorectal cancer (CRC). By integrating the strengths of both technologies, the researchers were able to capture not only gene-level expression but also alterations in transcript structures, such as alternative splicing and end processing events. This integration facilitated a more comprehensive understanding of the transcriptomic landscape and provided a detailed view of the diverse isoforms present in CRC.
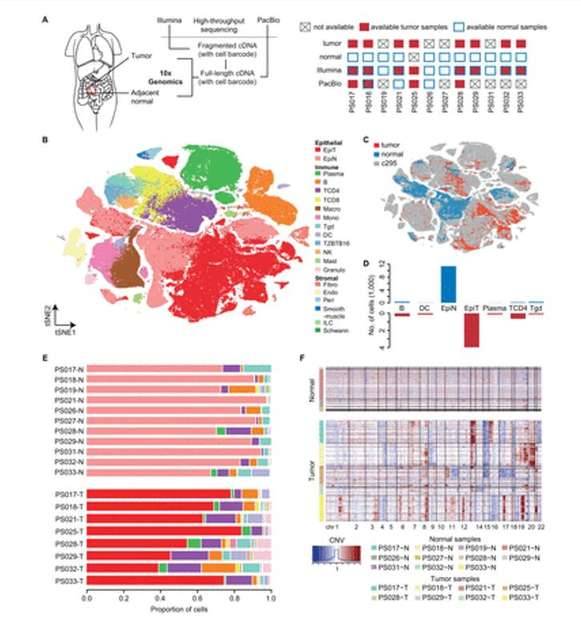 The short-read single-cell transcriptomic atlas of human CRC. (Li et al., 2023)
The short-read single-cell transcriptomic atlas of human CRC. (Li et al., 2023)
Unveiling Dysregulated Transcript Structures
The isoform-resolution transcriptomic atlas of CRC revealed a total of 394 dysregulated transcript structures within tumor epithelial cells. These structures included 299 instances resulting from various combinations of multiple splicing events. By characterizing and quantifying these dysregulated isoforms, the study shed light on the underlying mechanisms driving tumorigenesis and disease progression in CRC. These findings significantly enhance our understanding of the complex interplay between alternative splicing events and the development of CRC.
Characterization of Epithelial Lineages and Prognostic Subpopulations
Another crucial aspect of the study involved the characterization of genes and isoforms associated with specific epithelial lineages and subpopulations within CRC. By dissecting the molecular signatures of different subgroups, the researchers identified distinct prognoses among these subpopulations. This characterization not only elucidates the diverse genetic landscape of CRC but also provides valuable insights into the underlying mechanisms driving tumor heterogeneity. The knowledge gained from this characterization lays the foundation for developing tailored treatment strategies that target specific epithelial lineages, ultimately improving therapeutic efficacy and patient outcomes.
Discovery of Novel Neoepitopes
In addition to unraveling the heterogeneities within colorectal cancer (CRC), the study's integrated approach facilitated the identification of novel neoepitopes. By amalgamating predicted open reading frames (ORFs) derived from recurrent tumor-specific transcripts with mass spectrometry data, the researchers successfully discerned a repertoire of recurring neoepitopes. These neoepitopes, distinctive markers of tumor-specific antigens, exhibit robust potential as targets for immunotherapy. The capacity to recognize and target these neoepitopes ushers in promising prospects for the development of neoantigen-based cancer vaccines. By precisely directing therapeutic interventions toward tumor cells while minimizing undesired effects, these vaccines hold the potential to revolutionize the realm of immunotherapy and enhance patient outcomes in CRC.
Integration with PacBio Iso-Seq Technology
The integration of the isoform-resolution transcriptomic atlas of CRC with PacBio Iso-Seq technology signifies a notable leap forward in tumor neoantigen exploration and personalized immunotherapies. A key advantage of PacBio Iso-Seq technology lies in its capability to capture a broad range of isoforms, encompassing those resulting from alternative splicing events. By leveraging the extensive sequencing capabilities of PacBio's long-read technology, researchers can acquire a more comprehensive portrayal of the transcriptomic landscape, thereby uncovering dysregulated transcript structures and neoepitopes that may have been previously overlooked.
In summary, the integration of the isoform-resolution transcriptomic atlas of CRC with PacBio Iso-Seq technology significantly advances our understanding of tumor heterogeneity and neoantigen discovery. This integrated approach paves the way for the development of individualized immunotherapies that target specific dysregulated transcript structures and neoepitopes, ultimately improving patient outcomes and revolutionizing the treatment of colorectal cancer.
Reference:
-
Li, Zhongxiao, et al. "An isoform-resolution transcriptomic atlas of colorectal cancer from long-read single-cell sequencing." bioRxiv (2023): 2023-04.
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.


 Sample Submission Guidelines
Sample Submission Guidelines
 The short-read single-cell transcriptomic atlas of human CRC. (Li et al., 2023)
The short-read single-cell transcriptomic atlas of human CRC. (Li et al., 2023)