The auditory system, a distinctive mammalian sensory system, possesses remarkable biophysical properties. Hereditary deafness, a prevalent sensorineural disorder, is primarily a single-gene disease exhibiting substantial genetic heterogeneity, divided into nonsyndromic (80% of cases) and syndromic (20% of cases) groups. Presently, research has identified around 125 genes associated with nonsyndromic deafness and approximately 300 genes linked to syndromic deafness. Some of these causative genes play crucial roles in specific cochlear cell types.
The cochlear organ comprises a limited number of cells, particularly auditory hair cells, with less than 15,000 found in the human cochlea. This scarcity poses challenges in unraveling the molecular mechanisms governing its development and physiology. In recent years, advances in cochlear gene and cell therapies, along with in-depth transcriptomic studies, have led to the analysis of specific cochlear cell types. However, there is still a lack of a comprehensive standardized transcriptome profile encompassing the entire differentiation process from the sensory organ's inception to full maturation.
To address this gap, the study titled "Single-cell transcriptomic profiling of the mouse cochlea: An atlas for targeted therapies" was published in PNAS. Using a mouse model, the research team examined over 120,000 cells across three critical cochlear developmental stages: P8 (pre-hearing), P12 (during hearing), and P20 (cochlear formation). The objective was to create a comprehensive single-cell transcriptome profile of the mouse cochlea. By employing single-cell, mononuclear RNA sequencing (scRNAseq and snRNAseq) in combination with extensive RNA in situ hybridization assays (RNA scope), the transcriptomics of nearly all cochlear cell types were meticulously characterized. Remarkably, the researchers also uncovered three previously unknown cell types, a significant finding that lays the groundwork for deciphering the molecular mechanisms governing the unique organization of the basement membrane's biophysical properties.
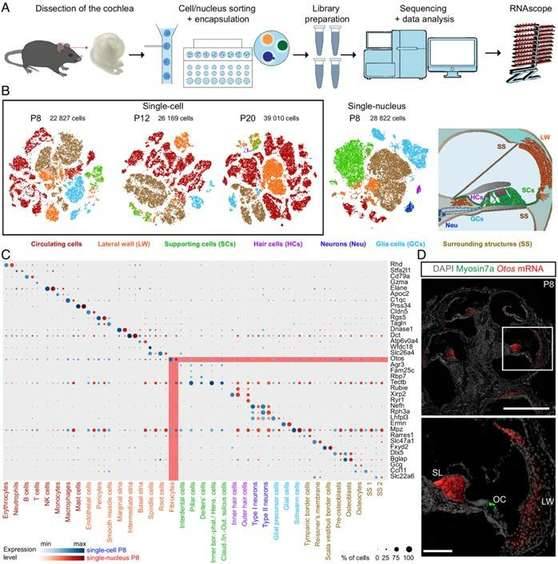 Transcriptomic characterization of cochlear cell types. (Jean et al., 2023)
Transcriptomic characterization of cochlear cell types. (Jean et al., 2023)
The team integrated single-cell transcriptomic data from three stages of differentiation. They divided the dataset into two main collections: circulating cells, mainly blood cells, and non-circulating cells. The non-circulating cells further differentiate into cells of soft and cartilaginous/osteochondral cochlear tissue. The circulating cells were characterized manually based on typical marker expression.
The non-circulating cell types were subdivided into six main groups according to their cochlear subregion of origin. These groups include the lateral wall cell group, the neurosensory epithelial cell group, and the spiral ganglion cell group. The remaining three cochlear cell types were grouped together as the "peripheral structure" cell group.
In their research, the team focused on cochlear cells expressing genes related to bone function and identified three different stages of maturation of osteoblasts. These stages were cochlear preosteoblasts, osteoblasts, and osteocytes. By analyzing the expression of Dlx5 and Runx2, which are involved in osteoblast differentiation, the researchers successfully identified these cell types. Among them, osteoblasts accounted for approximately 90%-95% of the total osteoblast population.
In addition to the three known cell types, the researchers discovered two previously unidentified cell clusters referred to as peripheral structures 1 and 2 cells (SS1, SS2). The SS1-specific expressed genes were Osr2 and Chrdl1, while the SS2-specific expressed genes were Bmp6 and Col1a1. The Gene Ontology (GO) analysis revealed that these genes play roles in osteoblast differentiation, ossification, cartilage development, odontogenesis, and biological processes related to skeletal system development.
In summary, apart from the three classical osteoblasts, the team unveiled two distinct cell types localized to spongy bone structures. These cells demonstrated osteoblast-like transcriptomic profiles but were different from pre-osteoblasts, osteoblasts, and osteocytes.
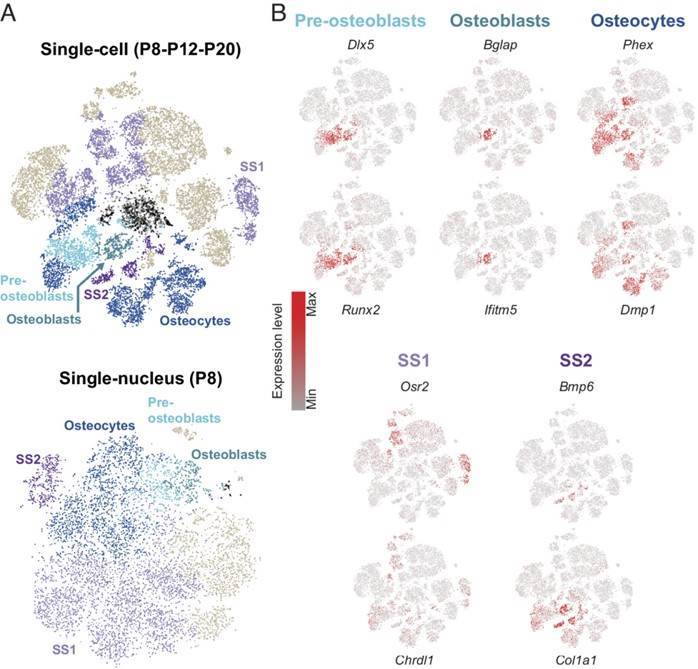 Transcriptomic characterization of the osseous cell types. (Jean et al., 2023)
Transcriptomic characterization of the osseous cell types. (Jean et al., 2023)
The research team examined alterations in gene expression within different cochlear cell types from P8 to P20 to evaluate their distinct and relative maturation processes. The findings revealed that during this developmental period, SVB cells, RM cells, and osteoblasts exhibited minor to moderate changes in gene expression. Conversely, cell types like fibroblasts, osteoblasts, SS1, and SS2 cells demonstrated substantial changes, while fibroblasts displayed the most remarkable shifts in gene expression.
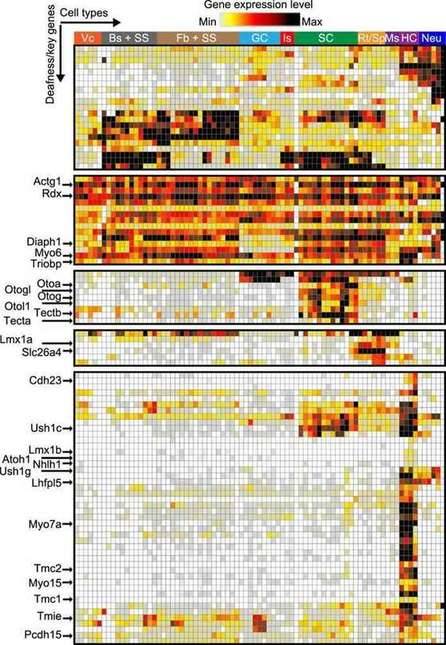 Cochlear cell expression pattern of nonsyndromic deafness genes. (Jean et al., 2023)
Cochlear cell expression pattern of nonsyndromic deafness genes. (Jean et al., 2023)
This study presents an extensive showcase of single-cell mapping throughout the process of cochlear development. It unveils the genes exhibiting the most significant variations in expression within cochlear cells at various time points during postnatal development. Moreover, the research offers in-depth transcriptomic data for multiple cochlear cell types, laying a strong groundwork for comprehending the origins of hereditary deafness.
Collectively, this comprehensive gene expression profile paves the path for unraveling the intricate gene regulatory network governing cochlear cell differentiation and maturation. Such insights are pivotal for devising effective targeted therapies aimed at fostering the development of potential treatments.
Reference:
-
Jean, Philippe, et al. "Single-cell transcriptomic profiling of the mouse cochlea: An atlas for targeted therapies." Proceedings of the National Academy of Sciences 120.26 (2023): e2221744120.
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.


 Sample Submission Guidelines
Sample Submission Guidelines
 Transcriptomic characterization of cochlear cell types. (Jean et al., 2023)
Transcriptomic characterization of cochlear cell types. (Jean et al., 2023) Transcriptomic characterization of the osseous cell types. (Jean et al., 2023)
Transcriptomic characterization of the osseous cell types. (Jean et al., 2023) Cochlear cell expression pattern of nonsyndromic deafness genes. (Jean et al., 2023)
Cochlear cell expression pattern of nonsyndromic deafness genes. (Jean et al., 2023)