The global challenge of antibiotic resistance (ARG) continues to grow, prompting the adoption of the 'One Health' approach, which seeks to optimize health for humans, animals, and the environment. Within this context, antibiotic-resistant bacteria can emerge in the gastrointestinal tracts of animals following antibiotic treatment, persisting within the animals and potentially posing a transmission risk to humans. Nonetheless, the degree of similarity in the acquired antibiotic resistance genes (ARGs) between humans and food animals, as well as the mechanisms driving the transfer of antibiotic resistance, remains shrouded in mystery.
This study delved into a vast and comprehensive dataset, comprising more than 1,000 intestinal samples sourced from humans, chickens, and pigs across various geographical locations worldwide. Additionally, they included 21 newly sequenced human gut microbiota samples for a more in-depth analysis. This extensive dataset served as the foundation for an investigation into mobile genetic element (MGE)-associated acquired antibiotic resistance genes (ARGs), opening novel pathways for comprehending the dynamics of antibiotic resistance gene transfer in food animals.
By selecting pigs and chickens as representative food animals, they compiled and examined a total of 1,487 metagenomes derived from human and food animal gut samples. This diverse dataset enabled us to emphasize the analysis of acquired antibiotic resistance genes (ARGs) linked to mobile genetic elements (MGEs) and to evaluate the transferability of ARGs by identifying the presence of MGEs and bacterial classifications in these two primary host types.
Diversity and Abundance of Antibiotic Resistance Genes (ARGs)
The analysis of metagenomic data revealed intriguing patterns in the diversity and relative abundance of ARGs across different host types. Most notably, the number of ARGs detected in human samples generally lagged behind those found in food animals, such as pigs and chickens. Furthermore, the beta diversity index exhibited greater variability in human samples compared to their counterparts from food animals. In particular, the relative abundance of ARGs was notably higher in pigs in comparison to human hosts.
Cluster analysis, which categorized ARGs based on their host origins, unveiled distinct patterns. ARGs related to tetracycline, vancomycin, and macrolide resistance were consistently widespread across the samples, transcending host distinctions. In contrast, quinolone, chloramphenicol, and aminoglycoside ARGs displayed significant variation among different hosts.
Remarkably, with the exception of cepA, encoding a class A β-lactamase, all ARGs exhibited higher levels of abundance in food animals as compared to humans.
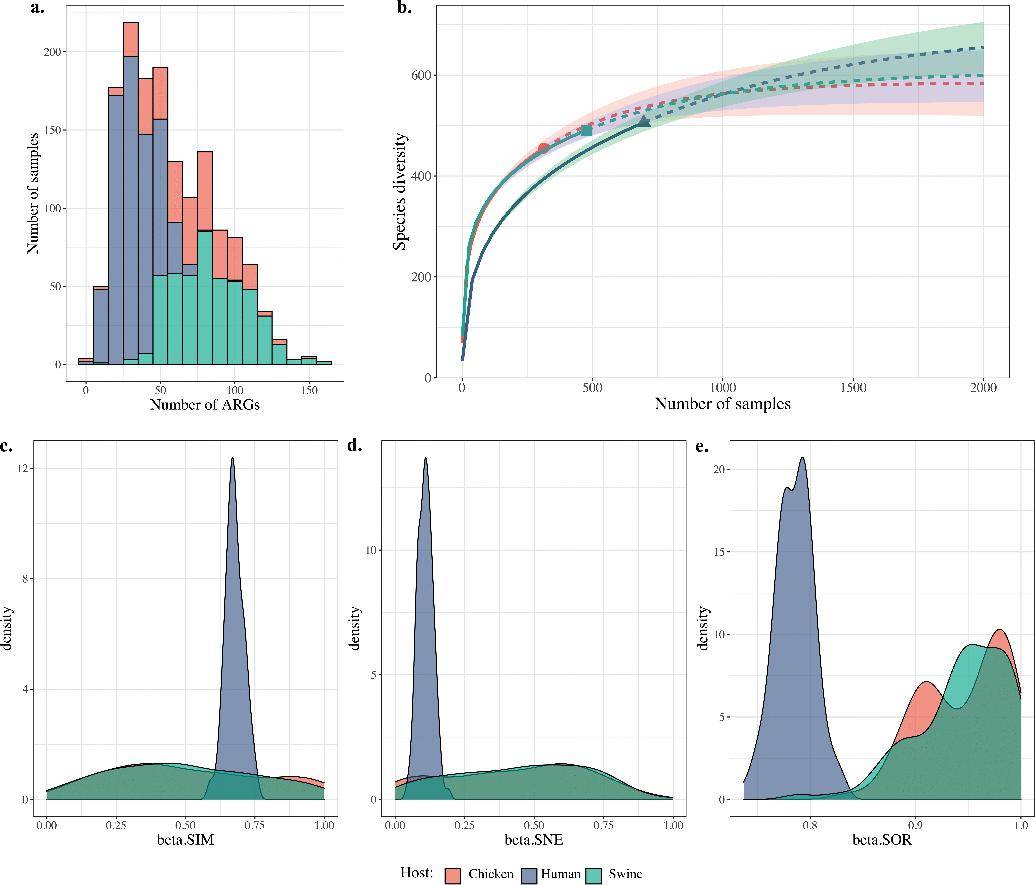 Diversity comparison of acquired ARGs in the hosts human, chicken, and swine. (Cao et al., 2022)
Diversity comparison of acquired ARGs in the hosts human, chicken, and swine. (Cao et al., 2022)
Shared Antibiotic Resistance Genes (ARGs) between Humans and Food Animals
Within this study, a total of 863 ARGs were identified, with 345 being common to both humans and pigs, and 214 shared between humans and chickens. Aside from ARGs resistant to clostridial acid, which were absent in human hosts, the occurrence of other ARG types was comparable across all hosts.
Exploring the potential transferability of these ARGs between the two host groups, the study identified insertion sequence (IS) elements in contigs containing ARGs in all samples. Symbiotic bacteria, particularly those of the Clostridium species, emerged as the primary source of mobile genetic element (MGE)-associated ARGs, found in both humans and food animals. Additionally, MGEs, such as Tn4451/Tn4453 and TnAs3, played a significant role in mediating ARG sharing between humans and food animals. TnpX, a member of the extensive dissociase family of site-specific recombinases facilitating the insertion of transient cyclic molecules, was also identified within Tn4451/Tn4453.
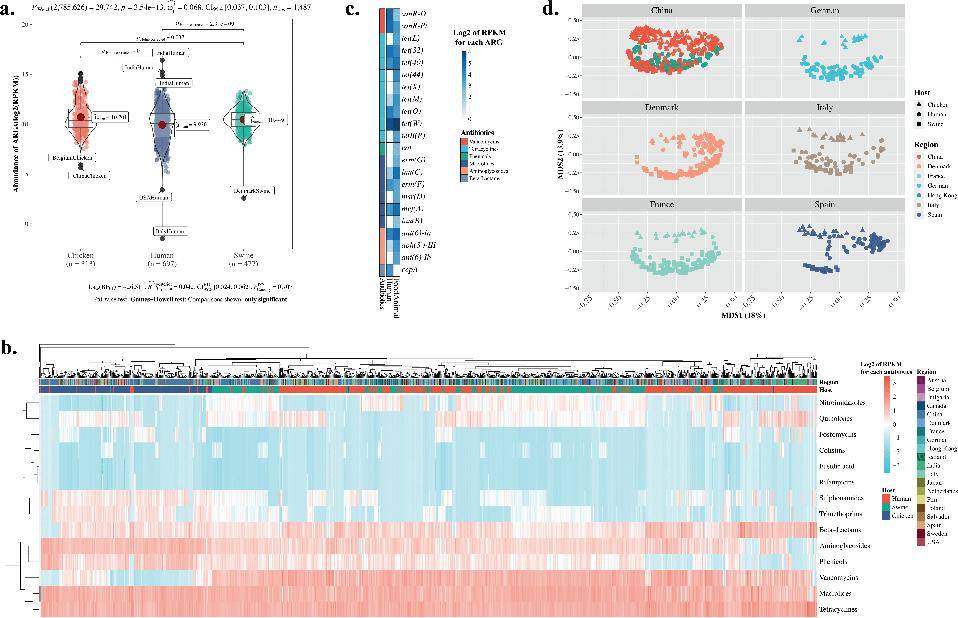 Comparison for the relative abundance of acquired ARGs in human and food animals. (Cao et al., 2022)
Comparison for the relative abundance of acquired ARGs in human and food animals. (Cao et al., 2022)
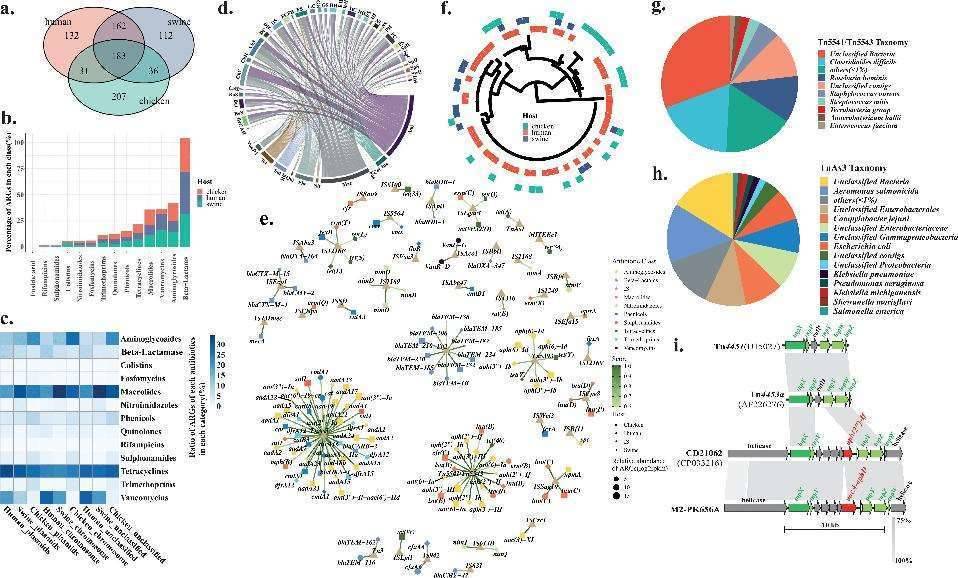 Shared ARGs in humans and food animals. (Cao et al., 2022)
Shared ARGs in humans and food animals. (Cao et al., 2022)
Transferability of ARGs in Human and Porcine Hosts and Key ARG Contributors
The stability of ARGs in both hosts was assessed using the Antibiotic Resistance Gene Transferability Index (ARGTI). Although the transferability of antibiotic resistance genes was more pronounced in humans than in pigs, the average transferability observed in pigs exceeded that in humans. Lefse analysis highlighted macrolide- and β-lactam-producing resistant ARGs as the principal contributors in both human and porcine hosts.
To summarize, this study unveiled potential antimicrobial resistance hotspots, particularly those ARGs with a high potential for horizontal transfer to pathogens. The discovery of these potential antibiotic resistance hotspots, particularly in food animals, underscores the imperative need for proactive monitoring to detect and mitigate emerging resistance threats. The findings from this study pave the way for an enhanced understanding of the stability and transfer of ARGs in food animals. Furthermore, this research holds promise for guiding the One Health initiative, thereby positively impacting human health.
Reference:
- Cao, Huiluo, et al. "Sharing of antimicrobial resistance genes between humans and food animals." MSystems 7.6 (2022): e00775-22.
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.


 Sample Submission Guidelines
Sample Submission Guidelines
 Diversity comparison of acquired ARGs in the hosts human, chicken, and swine. (Cao et al., 2022)
Diversity comparison of acquired ARGs in the hosts human, chicken, and swine. (Cao et al., 2022) Comparison for the relative abundance of acquired ARGs in human and food animals. (Cao et al., 2022)
Comparison for the relative abundance of acquired ARGs in human and food animals. (Cao et al., 2022) Shared ARGs in humans and food animals. (Cao et al., 2022)
Shared ARGs in humans and food animals. (Cao et al., 2022)