What is Repeat Sequence?
Repeat sequences play a crucial role in the complexity and functionality of genomes, and they can be broadly categorized into two types: scattered repeat sequences and tandem repeat sequences. Scattered repeats, such as transposons, are interspersed throughout the genome, while tandem repeats, like satellite DNA sequences, occur in a contiguous manner. This study delves into the analysis of repeat sequences within the genomes of approximately 600 insect species, shedding light on their diversity and distribution.
The Landscape of Repeat Sequences in Insect Genomes
The prevalence of repeat sequences in insect genomes is quite remarkable, ranging from 1.6% to a striking 81.5%. DNA transposons emerge as the most abundant type of repeat sequences, particularly prominent in the Coleoptera order, whereas Lepidoptera species exhibit a relatively lower proportion of DNA transposons.
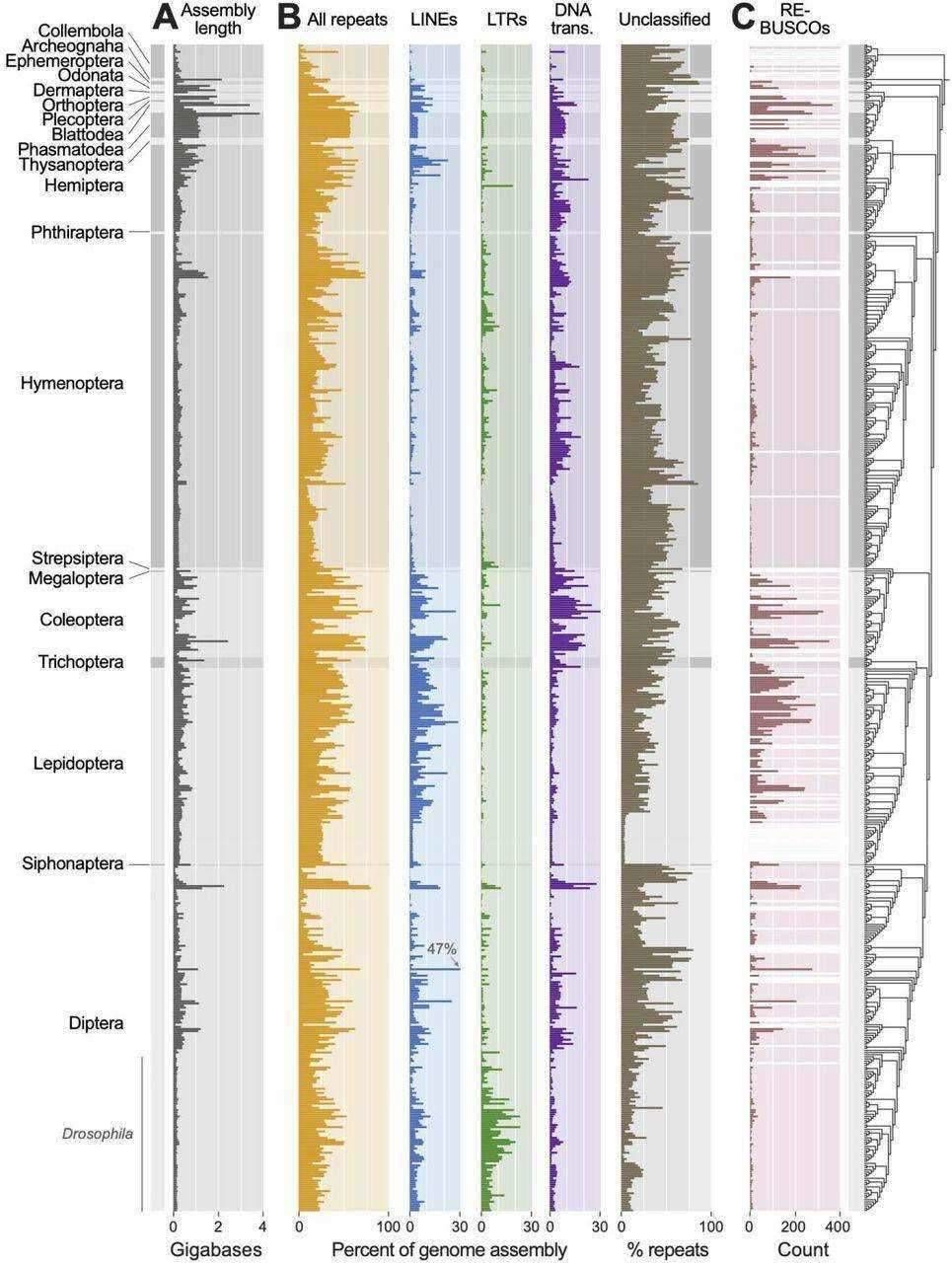 The repetitive element landscape of insects. (Sproul et al., 2023)
The repetitive element landscape of insects. (Sproul et al., 2023)
Line-Like Transposons - A Close Second
Line-like transposons, although the second most abundant repeat sequences, display substantial variation among insect orders. They are relatively less common in Hymenoptera, accounting for a mere 1.8% ± 1.7% of the genome.
LTR-Like Retrotransposons
In contrast, LTR-like retrotransposons are less widespread among insects but are noticeably abundant in Drosophila species. It's essential to acknowledge that the identification of LTR transposons can be challenging due to their larger and more intricate structure, possibly resulting in an underestimation of their prevalence in other insects.
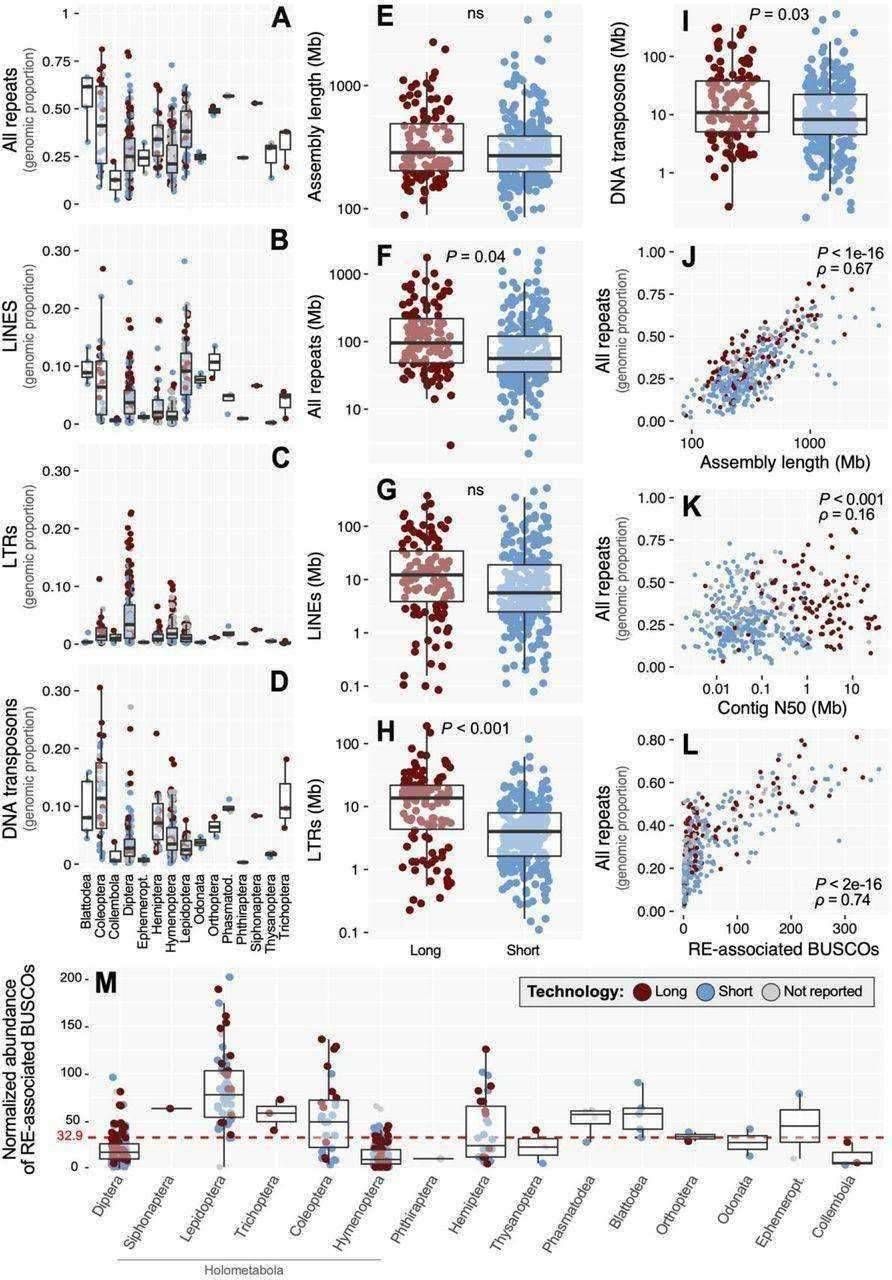 Statistical summaries of insect repetitive element dynamics, technology impacts. (Sproul et al., 2023)
Statistical summaries of insect repetitive element dynamics, technology impacts. (Sproul et al., 2023)
Genome Size Matters
Intriguingly, the study reveals a correlation between genome size and the presence of repeat sequences in insects. Larger genomes tend to harbor a greater abundance of these sequences.
Impact of Sequencing Methods
The investigation further assesses the effect of sequencing methods on the identification of repetitive sequences in genomes. Long-read sequencing outperforms short-read sequencing by identifying 36.1% more repeat sequences. This difference is most pronounced for LTRs, with a staggering 162% increase for long read lengths, followed by a 47% boost in the identification of DNA transposons.
Genes with Repetitive Sequences
Comparing genes that contain repetitive sequences (RE-associated BUSCOs), the study finds a positive association between the presence of LINE transposons and the number of such genes. For instance, in insects like Coleoptera and Hemiptera, these genes can constitute up to 25% of all genes, whereas Hymenoptera and Diptera species exhibit a much lower percentage, ranging from 1% to 2%.
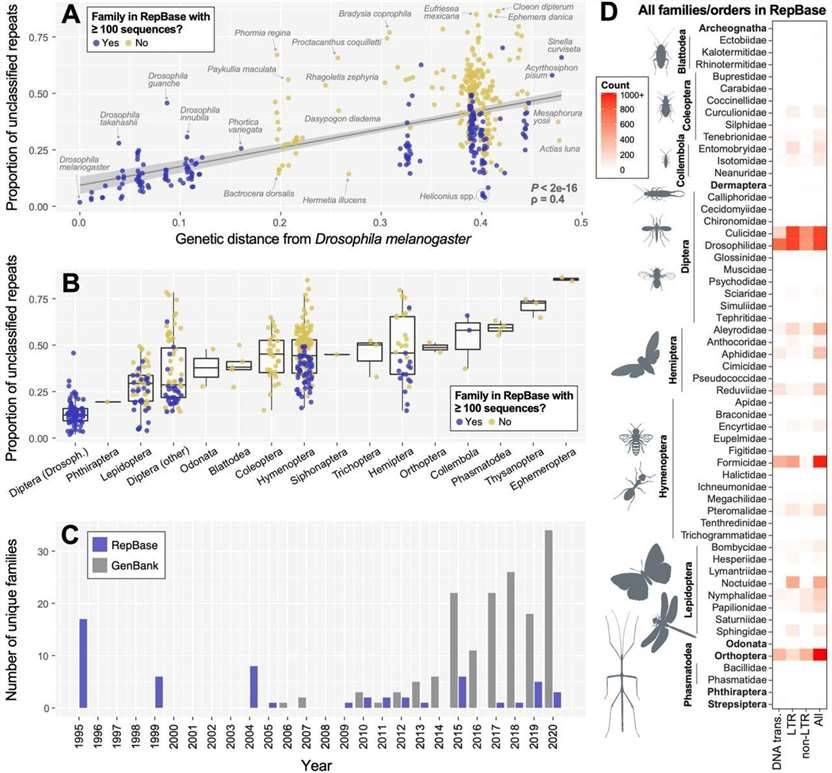 Insect representation in repetitive element databases and effects on RE detection. (Sproul et al., 2023)
Insect representation in repetitive element databases and effects on RE detection. (Sproul et al., 2023)
Impact of Reference Databases
Repetitive sequence identification heavily relies on reference databases, such as RepBase and Dfam. Notably, this study reveals that the divergence of insect species from Drosophila melanogaster is associated with a reduced ability to identify and classify repetitive sequences. For example, Drosophila species exhibit just 13.1% of unclassified repeats, while other insect classes may display as much as 40.5%. It's important to note that these unclassified repeats are generally shorter in length.
Database Bias
The predominance of repetitive sequences from mosquito and fruit fly families in the RepBase database underscores the significant impact of reference database bias on the annotation and identification of insect genomes.
Conclusion
This study provides a comprehensive overview of the abundance and distribution of repeat sequences in the genomes of various insect species. The findings emphasize the role of genome size, sequencing methods, and reference databases in shaping our understanding of repeat sequences. Furthermore, maintaining larger genomes with a higher abundance of repetitive sequences may confer adaptive advantages to certain insect species, as observed in the stone moth (caddisfly), where clades with larger genomes tend to exhibit higher polymorphism and occupy broader ecological niches.
Reference:
- Sproul, John, et al. "600+ insect genomes reveal repetitive element dynamics and highlight biodiversity-scale repeat annotation challenges." Genome Research (2023): gr-277387.
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.


 Sample Submission Guidelines
Sample Submission Guidelines
 The repetitive element landscape of insects. (Sproul et al., 2023)
The repetitive element landscape of insects. (Sproul et al., 2023) Statistical summaries of insect repetitive element dynamics, technology impacts. (Sproul et al., 2023)
Statistical summaries of insect repetitive element dynamics, technology impacts. (Sproul et al., 2023) Insect representation in repetitive element databases and effects on RE detection. (Sproul et al., 2023)
Insect representation in repetitive element databases and effects on RE detection. (Sproul et al., 2023)