Every year, an estimated 400,000 children around the globe are diagnosed with cancer. The peculiarities of pediatric cancers, when compared to their adult-onset counterparts, are not yet fully comprehended and cannot be entirely attributed to genetic factors. Transcriptome sequencing offers insights into the ongoing expression patterns of tumors, enabling tumor classification independent of the genome. Presently, most transcriptome-based classifiers are fully supervised tools that depend on existing markers within the tumor, limiting their ability to capture phenotypic variability. However, it is apparent that transcriptional variances exist within tumors, and even within the same tumor, these differences can yield both positive and negative prognostic indicators.
A Comprehensive Atlas of Pediatric Cancer
Nature Medicine recently published an article titled "Multiscale Transcriptomics Enables Diagnostic Classification of Childhood Cancer." In this study, the research team utilized a scale-adaptive clustering method called RACCOON, which employs RNA-seq for unsupervised classification of tumor subtypes. By applying RACCOON to a vast collection of 13,313 transcriptomes, they successfully constructed a comprehensive map of childhood cancer comprising 455 samples of both tumor and non-tumor origins.
The findings revealed that pediatric cancers exhibit a greater transcriptional diversity and maintain higher expression flexibility compared to adult cancers. Leveraging the insights gained from the RACCOON analysis, the researchers developed a novel integrated convolutional neural network classifier called OTTER. Remarkably, the results demonstrated that OTTER's predictions aligned with the clinicopathologic diagnosis of 82% of the patients in the prospective cohort. Moreover, OTTER provided additional clarity in diagnosing 7% of the cases that were previously ambiguous.
Overall, this study sheds light on the transcriptional specificity of childhood cancers and successfully validates a promising pan-cancer diagnostic tool.
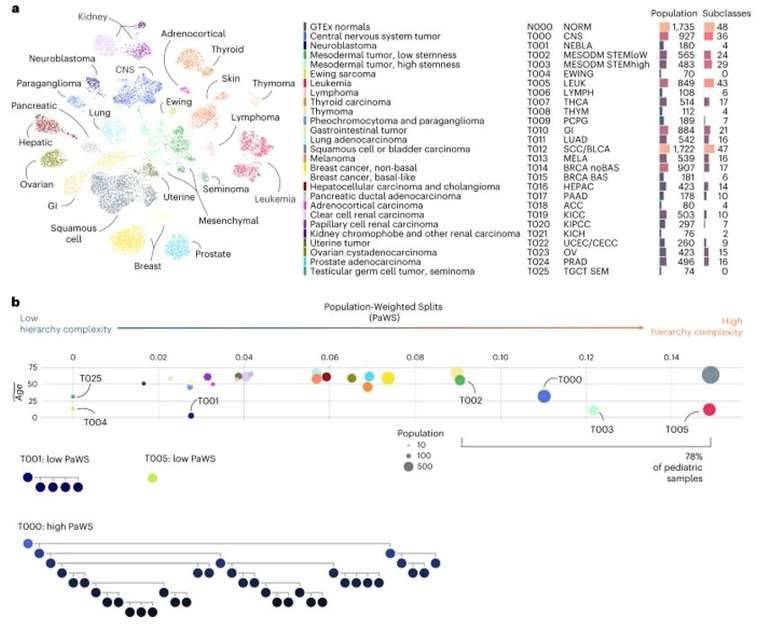 Transcriptional atlas of cancer. (Comitani et al., 2023)
Transcriptional atlas of cancer. (Comitani et al., 2023)
RACCOON: Delivering Accurate Cancer Classification
A breakthrough in childhood cancer research has been achieved by a team of scientists who developed a method to simplify the analysis of tumor transcriptome sequencing data and organize it into a hierarchical structure. This innovative approach allows for a more in-depth exploration of transcriptional variations both within and between tumor classes, leading to the discovery of new tumor subtypes. To accomplish this, the researchers harnessed the power of RACCOON, a cutting-edge technology that offers several performance advantages:
- Automatic Optimization: RACCOON automatically optimizes parameters for low-information filtering, downscaling, and clustering identification, minimizing the need for specialized knowledge in specific tumor types.
- Iterative Hierarchical Construction: RACCOON constructs hierarchies in a scale- and dataset-independent manner, providing a flexible and comprehensive framework for analysis.
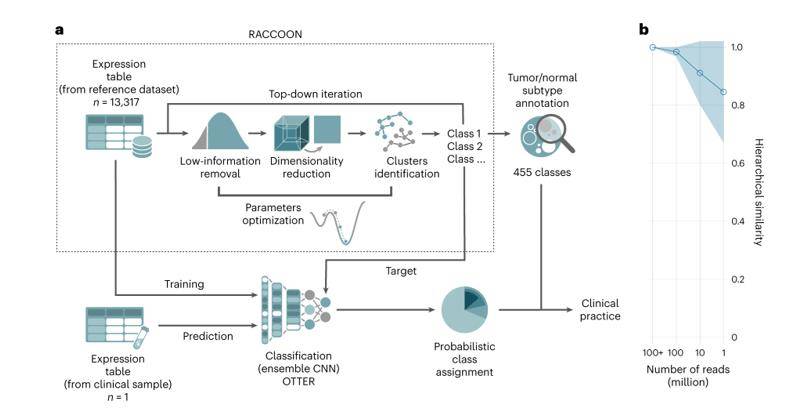 RNA-seq tumor subtype identification protocol. (Comitani et al., 2023)
RNA-seq tumor subtype identification protocol. (Comitani et al., 2023)
Building upon the RACCOON-generated hierarchies, the research team developed a state-of-the-art integrated CNN classifier called OTTER. OTTER demonstrates exceptional accuracy in matching individual patients to tumor classifications, surpassing the performance of any single model or previously published classifier. By accurately identifying multiple tumor and normal tissue components from clinical samples, OTTER accounts for the presence of various cell populations, mixed stromal or immune infiltrates, and offers a detailed depiction of tumor subtypes within a given tumor "lineage."
Notably, OTTER maintains its outstanding performance across all cancer types, even in the presence of multiple tumor mixtures, high normal contamination, technical noise, and shallow sequencing. Impressively, it achieves reliable tumor matching with only a few million reads, providing highly consistent predictions within minutes.
In summary, the implementation of RACCOON and OTTER has revolutionized cancer classification by providing precise and comprehensive insights into the molecular landscape of childhood cancers. This breakthrough technology has the potential to significantly advance our understanding of tumor subtypes and improve personalized treatment strategies for patients.
Pediatric Cancer Clustering Analysis using RACCOON
A novel clustering method, RACCOON, was employed for pediatric cancer analysis. This technique successfully categorized a total of 13,313 samples, including both tumor and non-tumor specimens, into 455 distinct categories across 8 levels. Within the highest level, 26 major tumor types were identified, and further subdivided into 48 subtypes. To facilitate comprehension of these classification structures, researchers introduced a scoring system named PaWS.
The PaWS scores revealed that four primary tumor types exhibited the highest values, constituting a significant portion of the entire sample cohort. Specifically, the pan leukemia group, squamous cell carcinoma, central nervous system tumors, and sarcomas collectively accounted for nearly 39% of all tumor samples. Interestingly, these four tumor types yielded a total of 192 tumor subtypes.
As anticipated, tumors originating from similar tissues tended to cluster together in the higher-level classification. However, the researchers made an intriguing observation regarding a new factor influencing transcriptional differences, namely age. A notable distinction in cancer types between adult and pediatric cases was evident. In fact, approximately 85% of pediatric cancers fell within just 6 out of the 26 high-level tumor types. Furthermore, many of these pediatric cancer subtypes represented novel discoveries in the field.
Reference:
-
Comitani, F., Nash, J.O., Cohen-Gogo, S. et al. Diagnostic classification of childhood cancer using multiscale transcriptomics. Nat Med 29, 656–666 (2023).
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.


 Sample Submission Guidelines
Sample Submission Guidelines
 Transcriptional atlas of cancer. (Comitani et al., 2023)
Transcriptional atlas of cancer. (Comitani et al., 2023) RNA-seq tumor subtype identification protocol. (Comitani et al., 2023)
RNA-seq tumor subtype identification protocol. (Comitani et al., 2023)