Background
Within the tumor genome, somatic mutations hold the ability to yield altered proteins that trigger immune responses against tumors. These mutated proteins fragment into peptides and are showcased on major histocompatibility complex (MHC) molecules. These distinct peptides, coined as "neoantigens," play a pivotal role in anti-tumor immune activity and have emerged as promising targets for therapeutic intervention. Although current investigations into neoantigens have predominantly concentrated on single nucleotide variants (SNVs) and insertion/deletion (Indel) mutations, the widespread structural variants (SVs) have been overlooked. SVs are pervasive in roughly 94.9% of tumors and possess the potential to generate neoantigens with diminished resemblance to the body's own structures, enhancing their immunogenicity. As a result, SVs stand as a significant wellspring of neoantigens. Nevertheless, a comprehensive analysis exploring the pan-cancer neoantigenic potential of SVs remains uncharted territory.
Through an examination of 2,528 neoantigens derived from structural variations across the genomes of 30 different cancer types, the investigators discovered a substantial and meaningful contribution of structural variations to the pool of neoantigens. This research highlights that the quantity and quality of mutation-induced neoantigens can more effectively capture the dynamics of interactions between tumors and the immune system compared to the sole consideration of tumor-specific neoantigens. These findings hold the potential to enhance the precision of patient selection for immunotherapeutic approaches. In essence, this study bridges a gap in the existing neoantigen knowledge base and serves as a valuable asset for the advancement of cancer vaccine development.
SV-Derived Neoantigens in Cancer
Utilizing the Neosv algorithm, the research team assessed 2,528 sets of tumor genomic data encompassing 30 distinct cancer types sourced from both the ICGC's Pan-Cancer Analysis of Whole Genomes (PCAWG) initiative and The Cancer Genome Atlas (TCGA). Their analysis revealed significant variation in the median count of neoantigens arising from structural variations (SVs) across the diverse cancer types. Notably, SV-derived neoantigens were notably concentrated in cancers exhibiting heightened chromosomal instability, notably including ovarian and breast cancers, esophageal adenocarcinoma, and gastric adenocarcinoma. Conversely, cancers characterized by relatively stable genomes, such as hematologic malignancies and most brain tumors (excluding glioblastoma), exhibited a scarcity of SV-derived neoantigens.
In addition, the researchers classified the SV-derived neoantigens based on their genomic and functional attributes, with the majority of neoantigen-generating SVs falling within the 1kb to 1Mb size range. As anticipated, SVs involving shifts in coding sequences accounted for a substantial 82.4% of the neoantigens. Furthermore, nearly 68% of the neoantigens were the result of unbalanced genomic events, including deletions and duplications. Of particular significance, the study identified that, apart from SVs spanning multiple genes, rearrangements involving a single gene – a facet often overlooked in gene fusion analyses – contributed to a noteworthy 41.2% of the novel antigens.
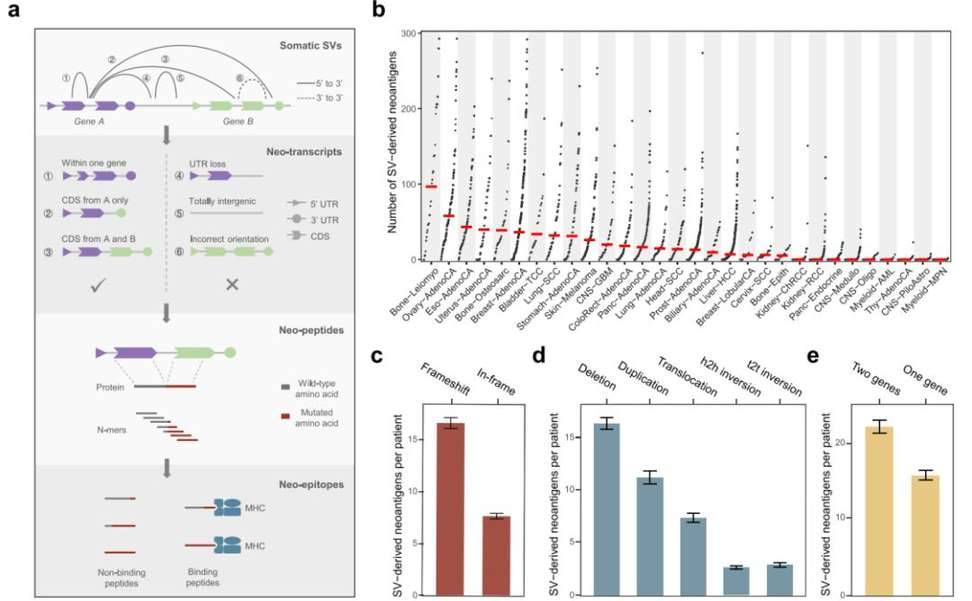 SV-derived neoantigens across 2528 tumors. (Shi et al., 2023)
SV-derived neoantigens across 2528 tumors. (Shi et al., 2023)
Structural Variants, SNVs, and Indels Exhibit Differential Impact
In addition to structural variants (SVs), single nucleotide variants (SNVs) and insertions/deletions (Indels) constitute two other primary genomic origins of neoantigens. By comparing these distinct types of neoantigens, the researchers observed a nearly exclusive distribution of neoantigens derived from SVs, SNVs, and Indels. In terms of quantity, the count of SV-generated neoantigens per patient was on par with Indel-derived neoantigens, yet notably lower than those originating from SNVs. This discrepancy in the neoantigen rates was attributed to the disruptive impact of SVs on open reading frames (ORFs), leading to a substantially higher rate of SV-derived neoantigens compared to SNVs and Indels.
It's worth highlighting that SVs emerged as the predominant neoantigen source in smooth bone leiomyosarcoma (comprising 61.6% of neoantigens) and osteosarcoma (constituting 49.1%). Additionally, SVs exerted significant influence in breast (37.7%) and ovarian (37.1%) cancers. These findings underscore the substantial role played by SVs in these specific cancer types, emphasizing their noteworthy contribution to neoantigen formation.
Subsequently, the scientists contrasted various epitope-associated measurements among SV, Indel, and SNV neoantigens. The findings indicated negligible variances in terms of binding affinity, binding stability, or hydrophobicity scores across mutation categories. Moreover, neoantigens derived from SVs exhibited a higher propensity for binding to T cells bearing diverse T cell receptors.
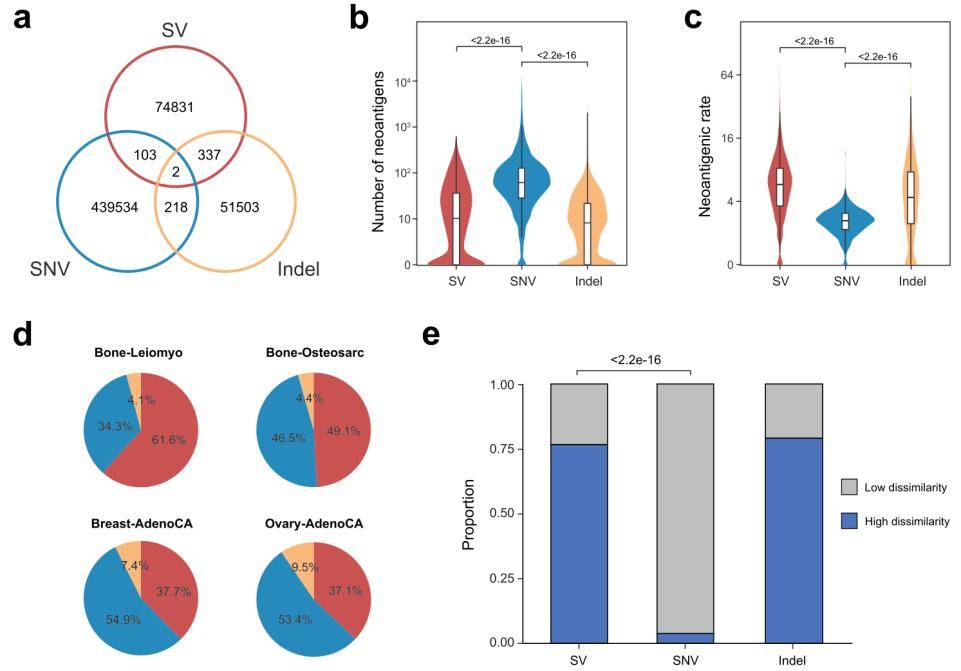 Comparison of the neoantigen repertoire derived from SVs to that from SNVs and indels. (Shi et al., 2023)
Comparison of the neoantigen repertoire derived from SVs to that from SNVs and indels. (Shi et al., 2023)
Reference:
- Shi, Yang, Biyang Jing, and Ruibin Xi. "Comprehensive analysis of neoantigens derived from structural variation across whole genomes from 2528 tumors." Genome Biology 24.1 (2023): 169.
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.


 Sample Submission Guidelines
Sample Submission Guidelines
 SV-derived neoantigens across 2528 tumors. (Shi et al., 2023)
SV-derived neoantigens across 2528 tumors. (Shi et al., 2023)  Comparison of the neoantigen repertoire derived from SVs to that from SNVs and indels. (Shi et al., 2023)
Comparison of the neoantigen repertoire derived from SVs to that from SNVs and indels. (Shi et al., 2023) 