HRD in Cancer Genomes
Cancer genomes serve as a record of genetic alterations that arise from DNA damage and defective DNA repair processes during the normal development of cells and the progression of carcinogenesis. These genetic alterations manifest as widespread somatic changes in BRCA1-deficient (BRCA1d) and BRCA2-deficient (BRCA2d) cancers, primarily linked to deficiencies in homologous recombination (HR), a pivotal pathway for repairing double-strand breaks (DSBs) in human cells. Some of these mutation patterns may represent specific error-prone mechanisms of DSB repair that cells employ in the absence of functional HR, potentially offering valuable biomarkers for identifying HR defects and guiding clinically relevant therapeutic strategies.
The compromised DSB repair in HR-deficient (HRD) cells is often associated with disruptions in structural genomic integrity, resulting in distinct cytogenetic abnormalities like radial chromosomes and chromosome bridges. Both microarray and whole genome sequencing (WGS) studies have revealed extensive patterns of heterozygous deletions (loss of heterozygosity or LOH) and other allelic imbalances in HRD cancers. It's worth noting that such copy number alterations can also be observed in tumors with intact HR (HRP) and are not tied to specific types of structural variants (SVs). Paradoxically, the distinguishing genomic features that set apart BRCA1d and BRCA2d tumors from HRP tumors include single nucleotide variants (SNVs), small deletions characterized by microhomology, tandem duplications, and simple deletions. Strikingly, these alterations have minimal impact on chromosome structure. The mechanisms underlying how aberrant DSB repair processes give rise to associated cytogenetic abnormalities in the presence of HR defects remain poorly understood.
Complex Structural Variations and Repair Mechanisms in BRCA1- and BRCA2-Deficient Cancers
Recently, an article was published in Nature. This study, titled 'Long-molecule scars of backup DNA repair in BRCA1- and BRCA2-deficient cancers,' harnessed the power of 46 deep strand-read whole genome sequencing (WGS) datasets from breast cancers with BRCA1 or BRCA2 mutations. Additionally, the team performed genome mapping analysis on short-read WGS data from thousands of tumors. This comprehensive approach unveiled a unique class of rearrangements, termed 'reversal pairs,' that are significantly enriched in HR-deficient cancers. The authors proposed a mechanism for HR-independent replication and restoration to account for the full spectrum of 'reversal pair' results. This discovery sheds light on a distinct category of rearrangements specific to BRCA1 or BRCA2 deficiencies, which underlie the cytogenetic abnormalities observed in HR-deficient cells.
To explore the intricate role of complex structural variations (SVs) in cancers associated with homologous recombination deficiency (HRD), the researchers in this study meticulously gathered data from 979 whole-genome sequencing profiles, primarily comprising primary tumors (95%). These tumors represented four distinct types frequently linked to HR defects. Among this dataset, there were 24 and 36 tumors displaying inactivation of both BRCA1 and BRCA2 alleles, alongside 487 tumors associated with HR pathway (HRP) abnormalities. Subsequently, the researchers employed an integrated genomic mapping approach to analyze copy number alterations and rearrangements, facilitating a comparative assessment of SV patterns between BRCA1-deficient (BRCA1d), BRCA2-deficient (BRCA2d), and HRP tumors.
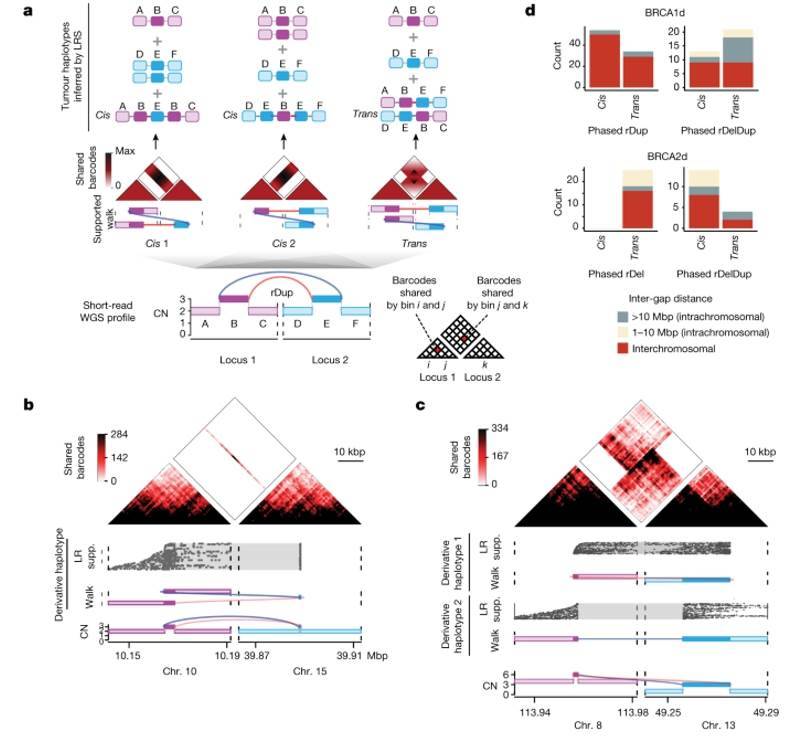 LR WGS reveals cis and trans phases for similar reciprocal pair topologies.
LR WGS reveals cis and trans phases for similar reciprocal pair topologies.
The findings unveiled remarkable similarities in SV profiles between HRP and HRD cancers, encompassing complex SVs. Nevertheless, a noteworthy exception emerged, as a particular class of SVs known as "Tandem Insertional Complexes" (TICs) displayed a significant enrichment in BRCA1d and BRCA2d tumors.
Classical reverse rearrangements, characterized by equilibrium translocations or inversions, are mechanisms that occur without any net loss or gain of genetic material, involving a pair of DNA junctions positioned adjacent to the broken ends within the same breakpoint. However, it is essential to note that many rearrangements, including translocations and inversions, deviate slightly from this classical definition, exhibiting near-reversal characteristics. These near-reversal SVs, such as TICs, entail copy number alterations within intermediate genomic regions referred to as "gap fragments." The polarity of these alterations is determined by the orientation of the joining of the broken ends. Specifically, copy gains result from connections between broken ends and gap fragments with a positive polarity (+), while copy losses occur when these connections involve the flanking segments with a negative polarity (-).
While TICs were notably enriched in BRCA1d and BRCA2d cancers, it is noteworthy that they were still discernible in a substantial proportion (36%) of HRP tumors. This suggests the possibility of uncovering distinctive HR-deficiency-specific patterns through a more comprehensive analysis of near-reverse junctions.
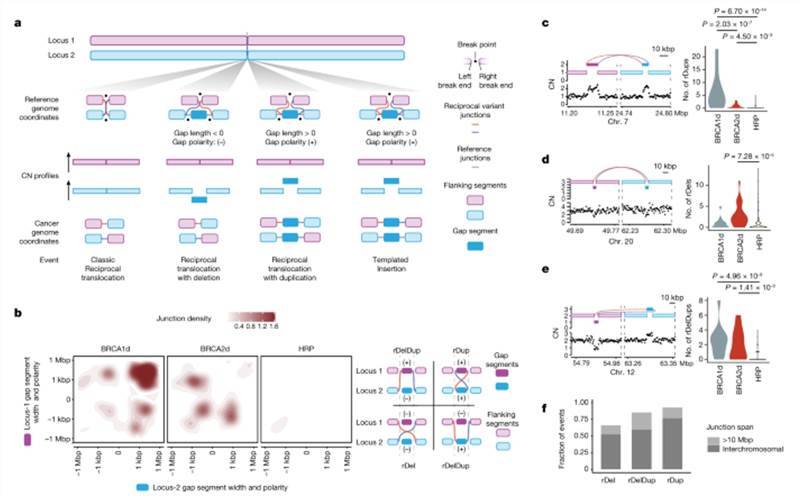 Reciprocal pairs are enriched in BRCA1d and BRCA2d tumors.
Reciprocal pairs are enriched in BRCA1d and BRCA2d tumors.
Based on these hypotheses, the researchers conducted a comprehensive analysis by comparing the structural variations (SVs) in close proximity across different genotypes. Their investigation revealed that paired circulating patterns were notably more abundant in BRCA1-deficient (BRCA1d) and BRCA2-deficient (BRCA2d) cancers when compared to high-risk population (HRP) cancers. These paired circulating patterns are referred to as "reverse pairs" or "reciprocal pairs."
Furthermore, when visualizing the length and polarity of gap fragments within these reverse pairs, the researchers identified three distinct subpatterns specific to BRCA1-deficient and/or BRCA2-deficient cancers:
- In BRCA1-deficient tumors (BRCA1d), they observed reverse pairs characterized by two gap fragments ranging from 100 base pairs to 100 kilobases in length, both having a positive polarity. These were termed "reverse duplicates" (rDups).
- In BRCA2-deficient tumors (BRCA2d), the researchers found reverse pairs featuring two gap fragments spanning from 1 base pair to 10 kilobases in length, both having a negative polarity. These were termed "reverse deletions" (rDel).
- Additionally, they identified reverse pairs containing gap fragments ranging from 1 base pair to 100 kilobases in length, with opposite polarities (positive and negative). These reverse pairs were enriched in both BRCA1d and BRCA2d cancers and were referred to as "reverse deletion-duplication" (rDelDup).
The researchers conducted linked-read (LR) and standard whole-genome sequencing (WGS) on 46 tumor samples, each paired with its corresponding normal sample. These samples exhibited genetic or somatic mutations in BRCA1 or BRCA2 genes. LR WGS offers comprehensive genome-wide coverage of both tumor and normal samples by employing barcoded short-read sequencing of long DNA molecules. These elongated DNA molecules play a crucial role in disentangling reverse pairs into phased somatic haplotypes, shedding light on the underlying mechanisms and outcomes of structural variations (SVs).
The results of this study demonstrated the consistency between LR WGS mapping and the short-read WGS data analysis previously described. Furthermore, LR WGS revealed a significant insight into reverse pairs with identical rearrangement orientations, producing two distinct chromosomal outcomes that can only be distinguished using long molecular data. The first outcome, known as "cis," involves the replication of a small DNA fragment transferred to a distant genomic location. The second outcome, referred to as "trans," represents a well-balanced translocation or a multi-megabase inversion characterized by substantial repeats (approximately 10kb) at each junction. Interestingly, the investigation revealed that trans-transversal pairs are prevalent in cases of rDup, rDel, and rDelDup, contributing to extensive rearrangements in BRCA1d and BRCA2d tumors. These trans-reverse pairs exert considerable chromosomal effects, potentially linked to cytogenetically observable abnormalities typically associated with BRCA1 and BRCA2 inactivation.
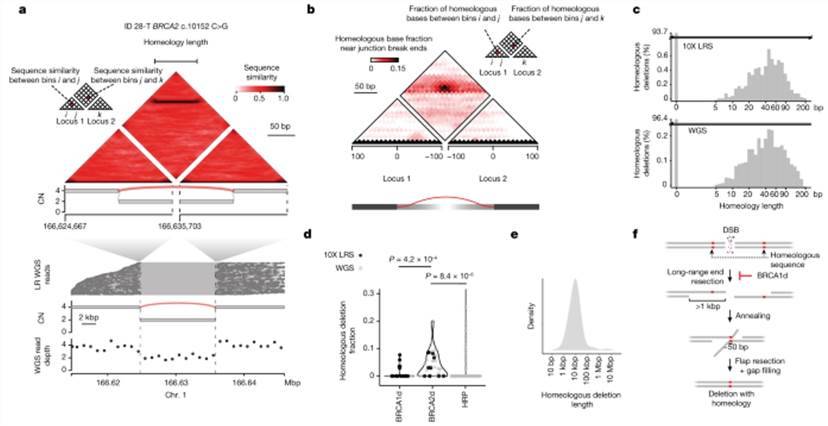 LR WGS uncovers footprints of SSA in BRCA2d genomes.
LR WGS uncovers footprints of SSA in BRCA2d genomes.
Differential Expression and Population-Specific Effects Potentially Driven by Genetic Factors
Utilizing eQTLs, our analysis revealed the identification of 9,150 eQTL loci associated with 5,198 distinct genes, of which 11% displayed ancestry-specific patterns. Among these, 812 eQTLs exhibited cell-type specificity, with 45% of them being predominantly detected in myeloid cells. Furthermore, we identified 1,505 viral-stimulated-response variants, known as reQTLs, linked to 1,213 genes. Notably, the genetic underpinnings of the myeloid response displayed viral specificity, with variations across cell populations.
Our investigation into inter-population differentially expressed genes and differential effect genes demonstrated that 56% and 60% of them, respectively, were associated with at least one eQTL. This suggests that the genes exhibiting the most significant population-based variation in differential expression and effects are likely influenced by genetic control, often linked to substantial effect-r loci. This highlights the presence of population differences in specific subsets of genes. For instance, between Africans and Europeans, we found that a single variant with strong population differentiation could explain 81-100% of the variation in GBP7 expression.
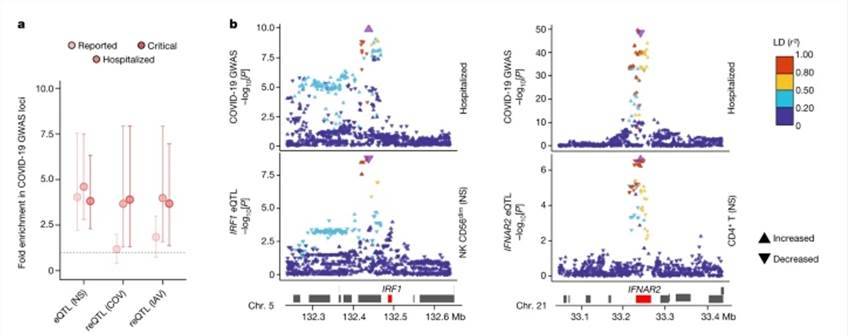 eQTLs and reQTLs contribute to COVID-19 risk. (Aquino et al., 2023)
eQTLs and reQTLs contribute to COVID-19 risk. (Aquino et al., 2023)
In-depth analysis further unveiled the presence of reverse SVs in both cis and trans orientations, each exhibiting specific topological and bundle length features correlating with replication restart mechanisms. Notably, these mechanisms encompass half-crossover and template insertion outcomes associated with break-induced replication (BIR). Based on these findings, a preliminary model is proposed, suggesting that microhomology-mediated BIR may explain the full spectrum of reverse pair mapping.
Additionally, LR WGS identified a distinct repair pathway known as single-stranded annealing (SSA) specific to BRCA2 defects in human cancers. This discovery highlights a unique strategy for reinforcing the repair pathway in cells with homologous recombination deficiency (HRD).
Reference:
-
Setton, Jeremy, et al. "Long-molecule scars of backup DNA repair in BRCA1-and BRCA2-deficient cancers." Nature (2023): 1-9.
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.


 Sample Submission Guidelines
Sample Submission Guidelines
 LR WGS reveals cis and trans phases for similar reciprocal pair topologies.
LR WGS reveals cis and trans phases for similar reciprocal pair topologies. Reciprocal pairs are enriched in BRCA1d and BRCA2d tumors.
Reciprocal pairs are enriched in BRCA1d and BRCA2d tumors. LR WGS uncovers footprints of SSA in BRCA2d genomes.
LR WGS uncovers footprints of SSA in BRCA2d genomes. eQTLs and reQTLs contribute to COVID-19 risk. (Aquino et al., 2023)
eQTLs and reQTLs contribute to COVID-19 risk. (Aquino et al., 2023)