According to statistics, cancer is the number one cause of death for patients under the age of 70. The earlier cancer is diagnosed and treated, the better the prognosis and survival rate of patients will be. Therefore, improving the efficiency and accuracy of early detection of cancer is crucial to the survival of cancer patients. In recent years, cell free DNA (cfDNA)-based non-invasive cancer screening technology has developed rapidly, and its application in the early detection and traceability of many cancers is promising.
Whole genome sequencing (WGS) is more sensitive in detecting low-load diseases compared to targeted deep sequencing. Recent studies have also demonstrated that screening cfDNA at the whole genome level is effective and feasible. Based on the original tumor map, ultra-sensitive monitoring of small residual disease can be performed using whole genome cumulative signals. However, this approach can only track the initial mutational profile of the patient's tumor tissue and cannot identify ab initio mutations. To date, cfDNA WGS has not been used for de novo cancer detection due to low confidence in cfDNA's ab initio mutation calling and inaccurate filtering. In addition, tumor-associated epigenomic features have not been fully explored across the genome and have not been used for cfDNA multi-tumor detection.
cfDNA Sequencing in Cancer Diagnosis
A recent paper entitled "Integrative modeling of tumor genomes and epigenomes for enhanced cancer diagnosis by cell-free DNA" uses advanced artificial intelligence (AI) algorithms to analyze the mutation density and pattern of cfDNA and epigenomes for superior accuracy in early cancer detection and histogenic localization.
The team first performed cfDNA whole genome sequencing, generating a training/validation dataset containing 3,366 samples at an average sequencing depth of 5× and 2.5× on MGI and Illumina sequencing platforms, respectively. The genomic model integrated large-scale reference cfDNA data from healthy cohorts and tumor tissue mutation data from the PCAWG project, using mutation distribution as model features; the epigenomic model integrated transposase-accessible and chromatin sequencing (ATAC-seq) based pan-cancer genome-wide chromatin mapping, using chromatin organization as model features. For the epigenomic model, the team identified tissue-specific NDRs (nucleosome deletion regions) by processing ATAC-seq data from 431 samples in public databases to analyze cancer type-specific cfDNA deletion patterns.
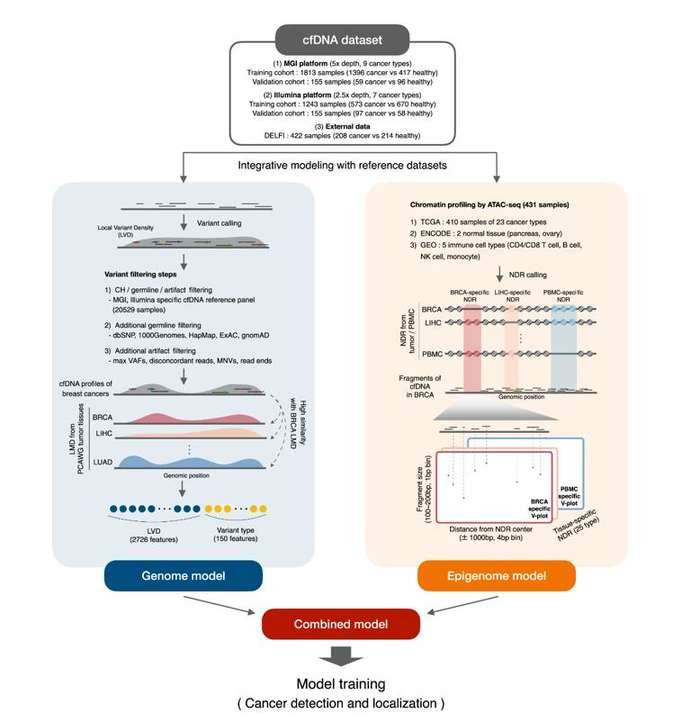 Integrative modeling of tumor genomes and epigenomes for enhanced cancer diagnosis by cell-free DNA. (Bae et al., 2023)
Integrative modeling of tumor genomes and epigenomes for enhanced cancer diagnosis by cell-free DNA. (Bae et al., 2023)
Conclusion
The team demonstrated the integration of a large-scale reference dataset to improve the sensitivity of cancer detection; secondly, the discovery of genomic and epigenomic features effective for cfDNA-based cancer diagnosis; and the construction of genomic models, epigenomic models, and combinatorial models based on this feature. The sensitivity of the combined model for detecting early-stage cancers (including pancreatic cancer) was comparable to that of late-stage cancers. This study investigated the relevance of these features to tumor biology from the perspective of genetic and epigenetic features of cancer, and laid the foundation for accurate cfDNA-based cancer diagnosis, especially in the early stages.
Improving Sensitivity through Reference Dataset Integration
To improve the sensitivity of cancer detection, the research team embarked on integrating a comprehensive reference dataset into their analysis. By incorporating a large-scale dataset, they expanded the scope of genetic and epigenetic features available for comparison and identification. This integration allowed for a more nuanced and accurate assessment of cfDNA profiles, boosting the sensitivity of cancer detection across various stages.
Unveiling Effective Genomic and Epigenomic Features
The study's breakthrough lies in the discovery of specific genomic and epigenomic features that prove effective for cfDNA-based cancer diagnosis. Through meticulous analysis, researchers identified genetic mutations and epigenetic modifications that were highly correlated with different types of cancers. These features provided valuable insights into the underlying tumor biology and served as crucial indicators for early-stage cancer detection, including notoriously challenging types like pancreatic cancer.
Construction of Comprehensive Models
Building upon the discovered genomic and epigenomic features, the research team constructed genomic models, epigenomic models, and combinatorial models. These models harnessed the power of machine learning algorithms to integrate and interpret the vast array of genetic and epigenetic data. By combining multiple features into comprehensive models, the researchers achieved remarkable sensitivity levels for both early-stage and late-stage cancer detection.
Relevance to Tumor Biology and Early-Stage Diagnosis
This study not only improves cancer detection accuracy but also highlights the significance of the identified features in understanding tumor biology. The genetic and epigenetic alterations uncovered through cfDNA analysis provide valuable insights into the molecular mechanisms underlying cancer development and progression. Furthermore, the study's emphasis on early-stage cancer detection demonstrates its potential to revolutionize diagnosis and improve patient outcomes by enabling interventions at the earliest possible stage.
Laying the Foundation for Precision Medicine
The integration of genomic and epigenomic features into cfDNA-based cancer diagnosis opens up new avenues for personalized medicine. By deciphering the unique genetic and epigenetic characteristics of individual tumors, clinicians can tailor treatment strategies to target specific vulnerabilities and bypass resistance mechanisms. This groundbreaking approach brings us closer to achieving precision medicine's ultimate goal: delivering targeted therapies that maximize efficacy while minimizing side effects.
Reference:
-
Bae, Mingyun, et al. "Integrative modeling of tumor genomes and epigenomes for enhanced cancer diagnosis by cell-free DNA." Nature Communications 14.1 (2023): 2017.
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.


 Sample Submission Guidelines
Sample Submission Guidelines
 Integrative modeling of tumor genomes and epigenomes for enhanced cancer diagnosis by cell-free DNA. (Bae et al., 2023)
Integrative modeling of tumor genomes and epigenomes for enhanced cancer diagnosis by cell-free DNA. (Bae et al., 2023)