We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy

Single nucleotide variants (SNVs) constitute approximately 58% of pathogenic genetic variations in humans, with 61% of these SNVs being of the "transition" type. As our understanding of how genomic DNA sequences impact human health has advanced, the potential of genomic editing technologies for clinical applications has become increasingly apparent. This progress has brought the ultimate human aspiration of precise gene editing closer to reality.
The CRISPR-Cas (clustered regularly interspaced short palindromic repeats and CRISPR-associated) system is a naturally occurring defense mechanism found in bacteria and archaea, which protects against viral invasions. Its widespread adoption in genomic editing is due to the programmable nature of its guide RNA and the robust activity of Cas nucleases across various cells and tissues. The CRISPR-Cas system achieves genomic editing by introducing double-strand DNA breaks (DSBs) at target sites within the cell. However, the DSBs induced by the CRISPR-Cas system can adversely affect genomic stability, necessitating the development of more reliable genomic editing technologies for precise DNA or RNA modifications.
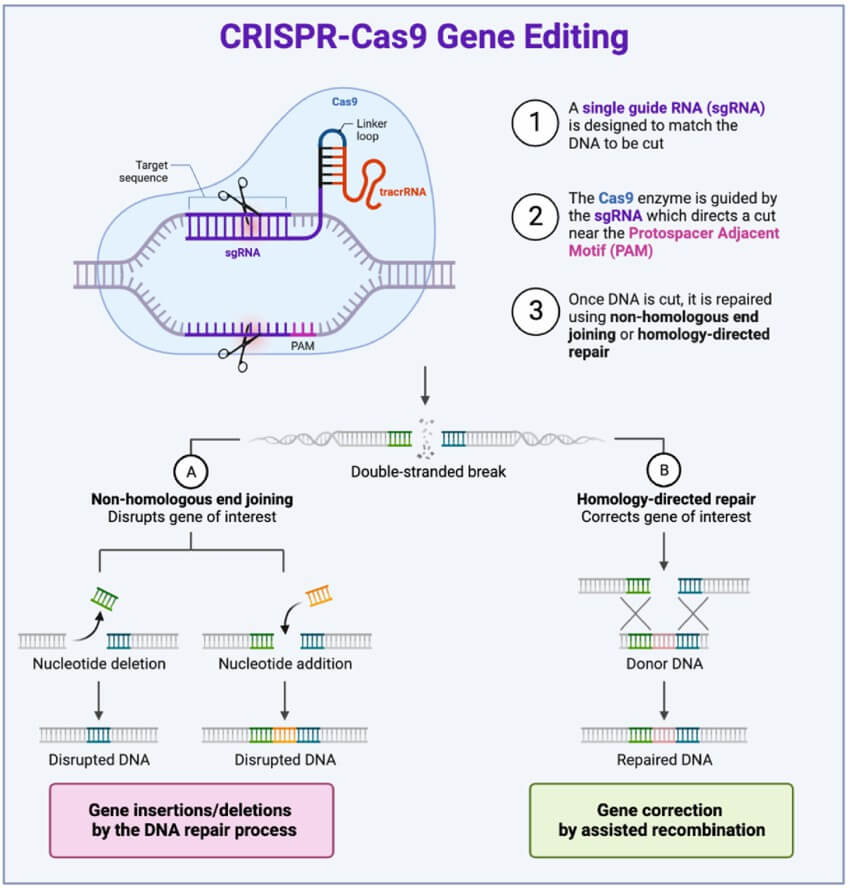 Figure 1. The process of CRISPR-Cas9 genome editing. (Arthur Yim et al,. 2024)
Figure 1. The process of CRISPR-Cas9 genome editing. (Arthur Yim et al,. 2024)
Since their inception in 2016, single-base editors (BEs) based on the CRISPR-Cas system have demonstrated substantial potential for clinical applications. Single-base editors are capable of precise correction of specific single nucleotide variants (SNVs) without introducing DSBs at the editing site, distinguishing them from Cas9, which generates DSBs. This characteristic renders BEs a safer genomic editing tool and provides promising solutions for the treatment of genetic disorders.
Mainstream BEs consist of a catalytically impaired Cas9 enzyme (nCas9) fused to a deaminase. Guided by a single-guide RNA (sgRNA), BEs target specific genomic locations. The nCas9 recognizes a sequence known as the protospacer adjacent motif (PAM), and the DNA segment targeted by the sgRNA is usually located upstream of the PAM. Complementary base pairing between sgRNA and the DNA sequence leads to the opening of the DNA double helix and the formation of an R-loop structure. The exposed non-target DNA strand then serves as a substrate for the deaminase. Within the deaminase editing window, the relevant type of base undergoes deamination, resulting in a base substitution, which is subsequently repaired through DNA repair mechanisms.
Based on the type of base substitution achieved by the deaminase, BEs are categorized into adenine base editors (ABEs) and cytosine base editors (CBEs). ABEs facilitate direct conversion of adenine to guanine, whereas CBEs convert cytosine to thymine. Collectively, ABEs and CBEs can address all types of SNV transitions. Clinically, these technologies have been utilized to correct pathogenic single nucleotide mutations in cell lines and murine models. For instance, they have been employed to ameliorate monogenic disorders such as β-thalassemia, β-thalassemia, and sickle cell anemia, as well as to restore visual function in adult mice suffering from Leber congenital amaurosis, a genetic retinal disease. Furthermore, the application of single-base editing systems in the ex vivo modification of chimeric antigen receptor (CAR)-T cells holds promising therapeutic potential in cancer immunotherapy.
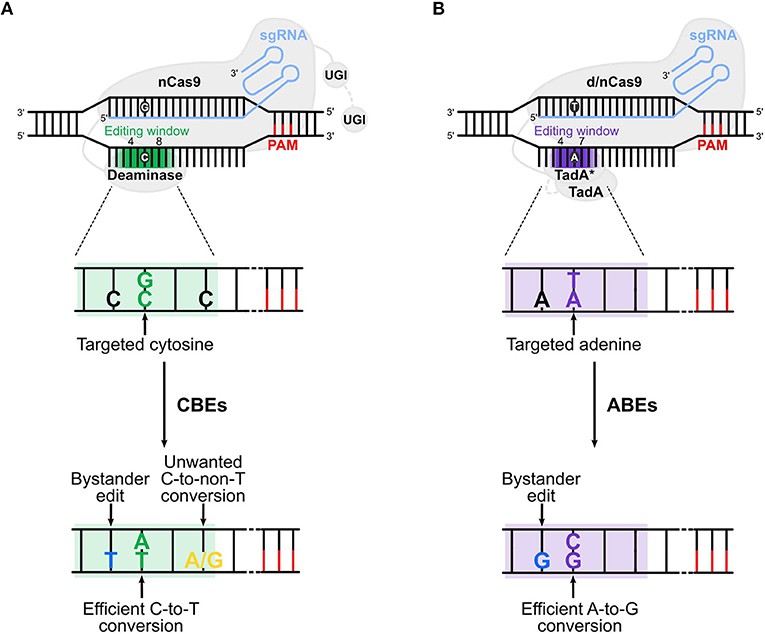 Figure 2. Schematic Representation of Gene Editing Mechanisms Employed by CBEs and ABEs (Panagiotis Antoniou et al,. 2021)
Figure 2. Schematic Representation of Gene Editing Mechanisms Employed by CBEs and ABEs (Panagiotis Antoniou et al,. 2021)
For gene editing tools to be effective therapeutic agents, they must exhibit high targeting efficiency and minimal harmful or off-target editing.
Existing research indicates that BEs may cause off-target edits at genomic sites similar to the targeted locus due to sequence similarity. This type of off-target editing, known as "sgRNA-dependent off-targeting," occurs because BEs can bind to genomic loci with high sequence homology to the target PAM. As a result, single-base editing can occur at these sgRNA-dependent off-target sites. In addition to sgRNA-dependent off-target editing, there is another form known as "sgRNA-independent off-targeting," which is introduced by the deaminase domain of BEs binding to DNA.
In 2019, researchers from the Yang Hui Laboratory at the Institute of Neuroscience, Chinese Academy of Sciences, and the Gao Caixia Laboratory at the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, reported that overexpression of the cytosine base editor BE3 in mouse embryos and rice resulted in random, genome-wide mutations, with average frequencies of 5×10−8/bp and 5.3×10−7/bp, respectively. These off-target edits may be attributed to the natural affinity of the deaminase domain of BE3 for DNA, independent of sgRNA-guided Cas9-DNA binding. Subsequent studies have demonstrated that CBEs can introduce sgRNA-independent off-target mutations in human induced pluripotent stem cells. Unlike sgRNA-dependent off-target editing, sgRNA-independent deamination occurs at various positions in the cell genome, making it challenging to identify off-target sites through targeted high-throughput sequencing. Comparisons of sgRNA-independent off-target activity between CBEs and ABEs have shown that CBEs exhibit a higher frequency of off-target events than ABEs. Modifying the deaminase domain or using alternative deaminases has yielded CBE variants with reduced sgRNA-independent off-target levels while maintaining high targeting efficiency (e.g., YE1-BE4, R33A-BE4, YE1-BE4-CP1028, YE1-BE4-NG, AmAPOBEC1, SsAPOBEC3B, YE1-BE3, FE1-BE3).
An alternative approach to achieve high targeting efficiency and low off-target effects involves delivering BEs as mRNA or ribonucleoprotein complexes into cells. This method reduces the duration of BE exposure within cells, thereby minimizing sgRNA-independent off-target editing. Similarly, delivering BEs as mRNA decreases sgRNA-independent off-target editing by shortening BE expression time. Thus, transient transfection of BE ribonucleoproteins or mRNA is advantageous over plasmid transfection and viral transduction in reducing off-target effects.
CD Genomics is dedicated to advancing research through comprehensive CRISPR/Cas9 off-target detection and analysis services.
Take the Next Step: Explore Related Services
Current methods for evaluating the specificity of CRISPR-Cas systems in eukaryotic cells fall into three main categories: computational tools (e.g., Cas-OFFinder, CFD, BEdeepoff), in vivo or in vitro capture of DNA double-strand break sites (e.g., DISCOVER-seq), and in vitro fragmentation of genomic DNA (e.g., Digenome-seq).
Computational methods predict potential off-target sites based on sequence similarity between the targeted and off-target sites. The latter two categories of methods are inadequate for assessing BE specificity in vivo, as BEs rarely induce double-strand breaks, which are necessary for these techniques. Instead, BEs primarily produce SNVs, which are not captured by these methods. To address these limitations, researchers have developed single-cell, single-clone WGS techniques. WGS enables the detection of SNVs and INDELs across the entire genome, identifying variations caused by gene editing tools by comparing edited genomes to control genomes. However, WGS is costly and challenging for high-throughput detection of BE off-target effects.
Recent advancements include the development of Detect-seq, a method for sensitive and efficient assessment of CBEs' off-target effects at the whole-genome level. Researchers exploit the intermediate product deoxyuridine (dU) generated by CBEs during editing. By biotinylating dU and enriching for it, and replacing normal C near dU sites with d5fC, Detect-seq generates a signal of consecutive C-to-T conversions through chemical labeling reactions, thereby enhancing the sensitivity of off-target detection. This technique has revealed two new types of sgRNA-dependent off-targets: out-of-protospacer edits and target-strand edits.
To mitigate sgRNA-independent off-target effects, the specificity of BEs can be enhanced by optimizing the deaminase domain. sgRNA-dependent off-targeting arises from the presence of similar sequences to the target site within the genome. Given that eukaryotic genomes contain numerous similar sequences to the target editing sequence, reducing sgRNA-dependent off-targeting remains a challenging issue. Currently, the only tool available for predicting sgRNA-dependent off-targeting of BEs is BEdeepoff. Prior to the development of BEdeepoff, predictions of BE off-target effects were generally based on tools designed for CRISPR/Cas systems.
Future research should explore the general principles governing the impact of sgRNA-DNA mismatches on off-target editing efficiency through additional single nucleotide BE experiments. At present, there remains a significant gap in the development of prediction tools specifically targeting off-target effects of BEs. The advancement of new high-throughput experimental methods for assessing BE off-target effects, such as Detect-seq, sgRNA-off-target site reporting systems, and high-precision techniques for detecting low-frequency mutations, presents new opportunities for efficient and cost-effective off-target evaluation.
References

CD Genomics is transforming biomedical potential into precision insights through seamless sequencing and advanced bioinformatics.
We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy