We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy

Next-Generation Sequencing (NGS) has caused a paradigm shift in the field of biological research since its inception. In comparison with traditional Sanger sequencing, NGS has been shown to significantly enhance the rate of data generation. Nevertheless, NGS necessitates the use of a greater volume of biological samples and is more costly, which hinders its application on a large scale and in batches, consequently limiting its utilisation in disease cohort analysis and drug screening.
In DRUG-seq for miniaturised high-throughput transcriptome profiling in drug discovery, researchers have for the first time combined the depth of barcode technology with second-generation sequencing for high-throughput drug screening. This breakthrough technology enables the simultaneous observation of tens of thousands of genes' transcriptome expression changes in drug screening, and it dramatically improves the screening efficiency and data dimension. In comparison with conventional drug screening methodologies, this technology successfully overcomes the limitations associated with extensive sample sizes, concurrently generating substantial quantities of transcriptome data. This facilitates the acquisition of comprehensive information regarding the molecular mechanisms underlying each drug candidate.
In order to enhance the adaptability of drug screening to batch transcriptome and to expand the application scenarios of DRUG-seq technology, DRUG-seq2 was developed.
DRUG-seq2 technology has been shown to exhibit a high level of efficiency that is comparable to that of second-generation sequencing. Furthermore, it has been demonstrated to offer a series of significant advantages that give it a wide range of potential for research and clinical applications.
Firstly, DRUG-seq2 has been shown to significantly reduce the amount of sample required. In comparison with traditional RNA-seq methods, DRUG-seq2 requires only a minute amount of sample, such as a single organoid, a small amount of blood or a tissue section, for large-scale sequencing. This feature renders it especially well-suited for large quantities of valuable samples that are difficult to obtain in a clinical context, such as tumour tissues or rare cell populations, where the minimum sample size can be as low as 1,000 cells/sample, greatly expanding the scope of the study.
Secondly, DRUG-seq2 technology has been demonstrated to simplify the experimental process by omitting the RNA extraction step in traditional RNA-seq and processing the cell samples directly. The removal of the complex step of RNA extraction has been shown to improve the efficiency of the experiment, reduce the variability of the experimental operation, shorten the experimental cycle, and reduce the workload of researchers.
Furthermore, DRUG-seq2 boasts significant advantages in terms of high-throughput and low-cost capabilities. This methodology facilitates the simultaneous sequencing of thousands of samples within a single experiment, thereby markedly enhancing the overall throughput and efficiency of the process. Furthermore, DRUG-seq2 offers a substantial cost advantage over traditional RNA-seq methods, with the cost of sequencing a single sample as low as $50. This renders it especially well-suited for parallel analysis of large-scale samples, providing an economical and feasible solution for efficient data collection.
DRUG-seq2 technology is distinguished by its remarkable flexibility in application, rendering it highly versatile across a wide spectrum of research domains, ranging from oncology research to drug development. Whether in the context of elucidating drug mechanisms or identifying tumour markers, DRUG-seq2 provides researchers with substantial genomics data support, thus establishing itself as an indispensable and significant instrument within both scientific research and clinical practice. In conclusion, DRUG-seq2 is a revolutionary technology in the field of genomics research, offering significant advantages such as ultra-low sample requirements, a simplified operational process, high throughput, and low cost, along with extensive application prospects.
Take the Next Step: Explore Related Services
Recent advancements in molecular technology, particularly DRUG-seq2, have had a profound impact on the fields of disease diagnosis, treatment and drug discovery. DRUG-seq2 facilitates precise analysis of gene expression in drug-treated cells, thereby contributing to the advancement of knowledge in the areas of disease mechanisms, tumour traceability and drug responses. It has demonstrated efficacy in the tracking of viral persistence, including SARS-CoV-2, and its association with long-term symptoms, while also enhancing tumour diagnosis and personalised treatments. In the field of drug discovery, DRUG-seq2 facilitates the identification of drug targets and toxicity risks, thereby supporting the modernisation of traditional Chinese medicine. Furthermore, DRUG-seq2 optimises cell culture conditions, thereby facilitating the development of improved models for drug screening and disease research, and thus ultimately advancing medical care and drug development.
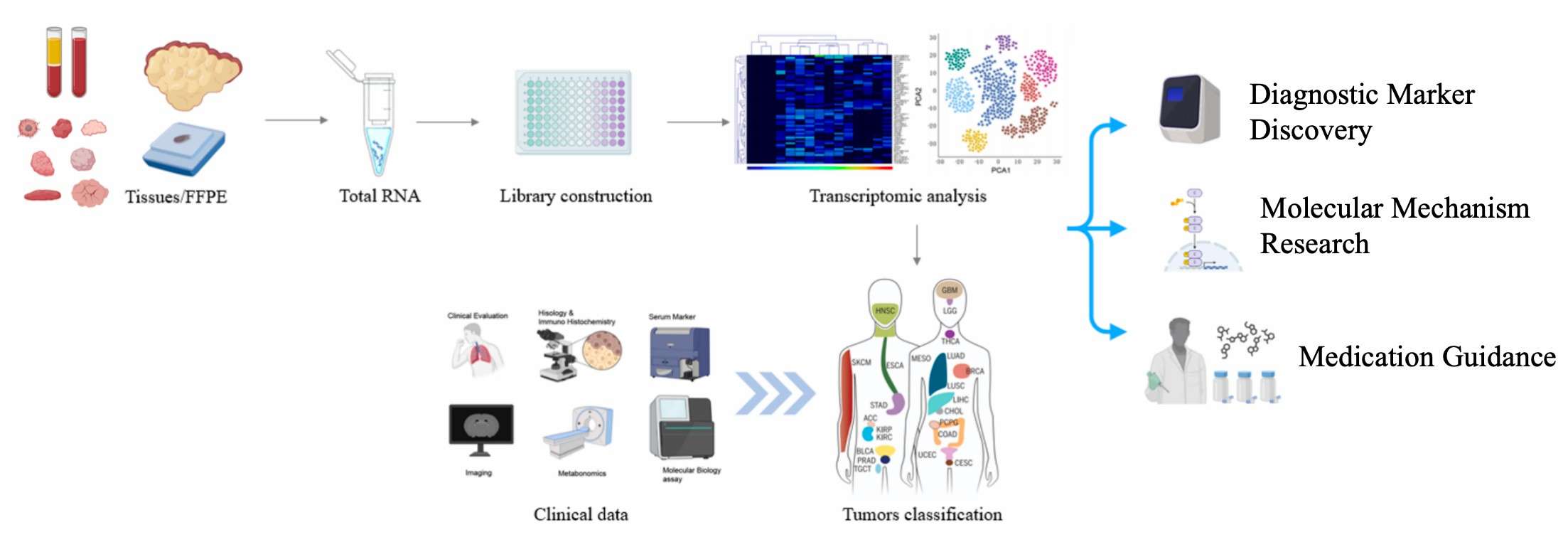
In recent years, molecular technology has undergone continuous development, with the result that precise diagnosis and treatment of diseases have become a major focus of medical research. DRUG-seq2 technology enables researchers to provide more precise and detailed data support in the diagnosis, traceability and treatment of a wide range of diseases.DRUG-seq2 technology can not only reveal the characteristics of diseases and tumours, but also help to discover potential drug therapy markers, thus providing a new direction for clinical treatment.
 Discovery of molecular signatures of various tissues in diseased/sub-healthy populations using DRUG-seq2 (Zuo et al., 2024)
Discovery of molecular signatures of various tissues in diseased/sub-healthy populations using DRUG-seq2 (Zuo et al., 2024)
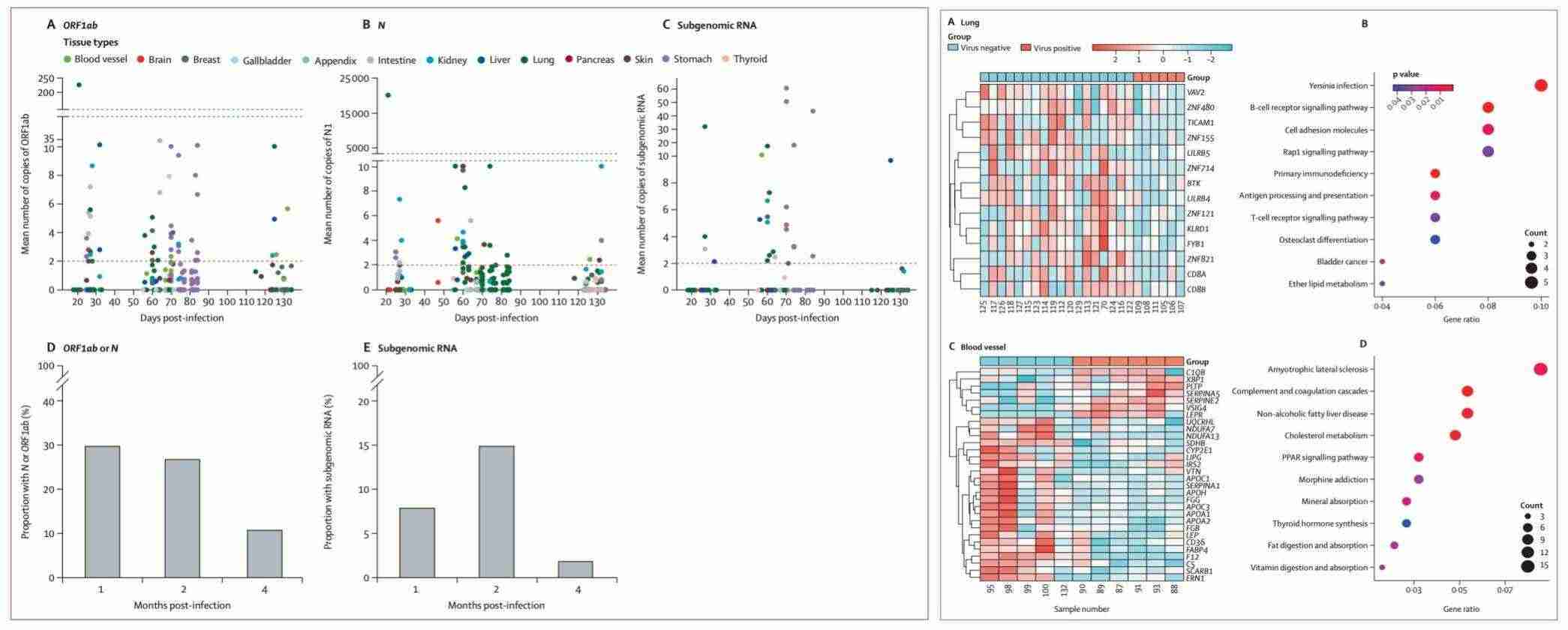
Firstly, researchers conducted in-depth analyses of tissue samples from neocoronavirus-infected patients using the DRUG-seq2 technique. These analyses revealed that viral RNA persisted in multiple tissues for a protracted period of time, and that this persistence was strongly associated with the development of long-term symptoms of the disease. Utilising a cross-sectional cohort study of Chinese patients infected with the new coronavirus, the researchers identified that viral RNA persisted in various tissues in the patients' bodies from 1 to 4 months following infection. Furthermore, the persistence of this viral RNA was found to be associated with long-term symptoms, such as fatigue, in the patients. The results of this study suggest that persistent SARS-CoV-2 infection may be a potential causative factor for long-term symptoms, providing new ideas for the treatment of neocoronary sequelae.
 Tumour tissue traceability and its diagnostic marker development using DRUG-seq2
Tumour tissue traceability and its diagnostic marker development using DRUG-seq2
Secondly, DRUG-seq2 technology plays a pivotal role in tumour traceability, as the transcriptional profiles of tumour cells reflect their biological characteristics and can reveal the origin of their foci of origin. This is crucial for tumour diagnosis and treatment. By analysing the specific gene expression patterns of tumour cells, doctors can accurately determine the possible site of origin of a tumour and thus provide more precise treatment plans for patients. Furthermore, the transcriptional profiling data facilitates not only the identification of the tumour's origin but also the personalisation of treatment and the prediction of patient response to specific drugs, thereby enhancing the targeting and effectiveness of treatment. In light of the advancements in targeted therapy and immunotherapy, it is anticipated that this transcriptional profiling-based technology will assume an increasingly pivotal role in the management of tumours.
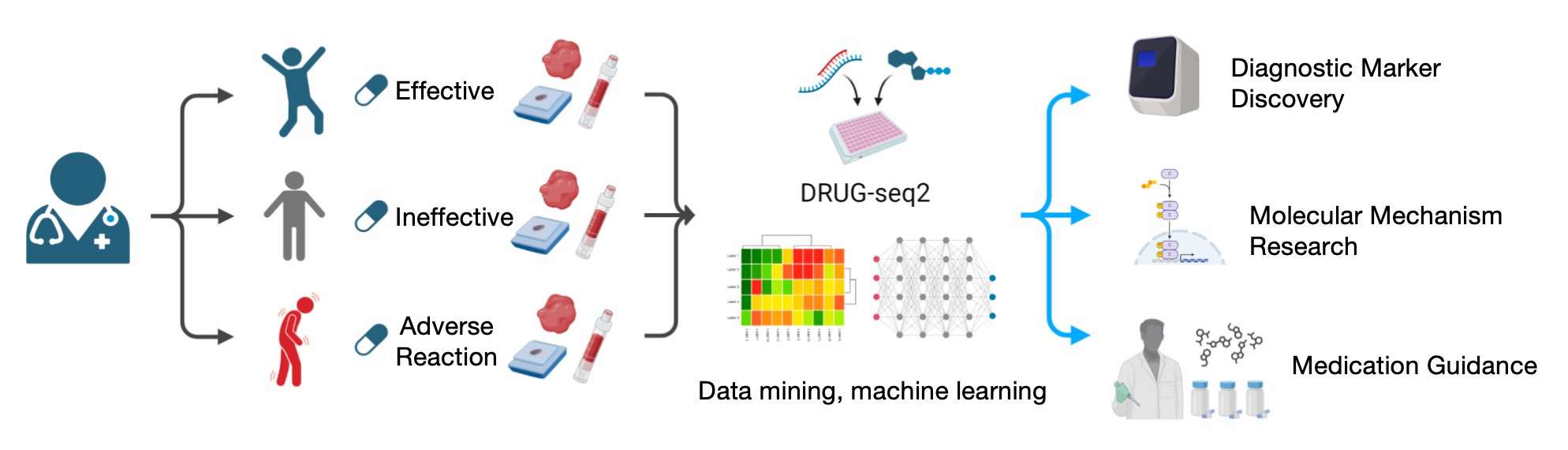 Discovery of diagnostic markers and their molecular mechanisms in drug-sensitive populations using DRUG-seq2
Discovery of diagnostic markers and their molecular mechanisms in drug-sensitive populations using DRUG-seq2
Furthermore, DRUG-seq2 technology has the capacity to identify diagnostic markers and their molecular mechanisms in drug-sensitive populations. By collecting tissue samples from patients in bulk and sequencing them efficiently, combined with machine learning and artificial intelligence models, researchers can analyse patient populations with different treatment prognoses and identify potential markers for predicting the effects of drug therapy. These markers can not only help predict a patient's response to a specific drug, but also provide clues to new mechanisms of drug action. The integration of multimodal data, alongside AI techniques such as the MuMo model, facilitates a comprehensive capture of patients' disease characteristics and an effective management of missing data, thereby enhancing the accuracy of clinical decision-making.
In conclusion, the application of DRUG-seq2 technology provides powerful technical support for the precise diagnosis of diseases, the traceability of tumours and their treatment, and the personalised protocols of drug therapy. By combining artificial intelligence and machine learning technologies, future medical research and clinical practice will be able to achieve more efficient and precise treatments. The continuous development of these technologies is expected to further advance the frontiers of medicine, thereby offering patients greater hope for treatment and the possibility of improving their quality of life.
In the domain of drug discovery, precise evaluation of the effects of drug candidates and their molecular mechanisms is a pivotal step in accelerating the process of discovering new drugs. The emergence of DRUG-seq2 technology provides a novel perspective on this process by examining the transcriptome of drug-treated cells, thereby revealing the drug's pathways of action, gene expression changes, and cellular responses. This multi-dimensional analysis facilitates researchers in identifying potential targets and toxicity risks of drugs, in addition to predicting the long-term effects and side effects of drugs. Consequently, DRUG-seq2 provides more precise guidance for the clinical application of drugs and significantly improves the efficiency and accuracy of drug development.
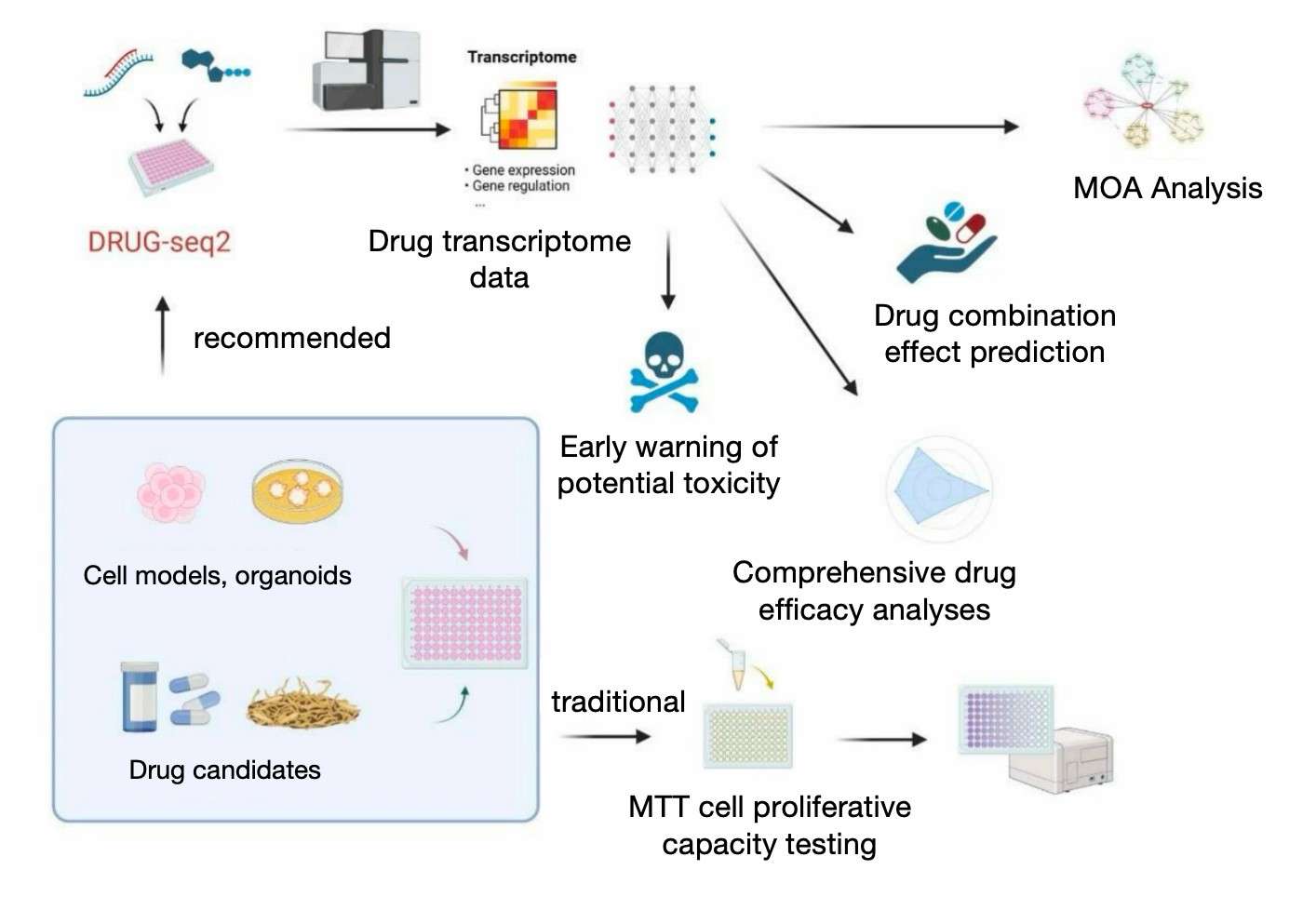 Comprehensive evaluation of the effects and molecular mechanisms of drug candidates using DRUG-seq2
Comprehensive evaluation of the effects and molecular mechanisms of drug candidates using DRUG-seq2
The employment of DRUG-seq2 technology in the realm of new drug discovery offers a comprehensive perspective on gene expression and drug response, facilitating the evaluation of drug candidates' molecular mechanisms. The technology's primary strength lies in its capacity to elucidate the biological processes influenced by drugs and to further refine the mechanisms of drug action through the analysis of transcriptional profiles from multiple dimensions. This technology enables researchers to not only swiftly obtain an assessment of the initial effects of a drug, but also to identify possible side effects from an early stage, which is crucial for the long-term clinical application of the drug.In particular, in preclinical studies of drug candidates, DRUG-seq2 can provide substantial data support for the evaluation of drug safety and efficacy.
DRUG-seq2 has also been identified as a valuable tool for the advancement of research in the field of Chinese medicine. Through the analysis of the transcriptome of traditional Chinese medicine (TCM) monomers or components, researchers are able to elucidate alterations in gene expression across diverse cell and tissue models. This, in turn, facilitates a more profound comprehension of the mechanisms by which these components impact biological pathways and disease processes. The technology facilitates the verification of the known pharmacological mechanisms of TCM components and the potential discovery of their roles in new indications or new biological targets.DRUG-seq2's high-throughput sequencing capability and precise data analysis provide significant support for the modernisation of TCM and propel research in this field towards the era of histology.
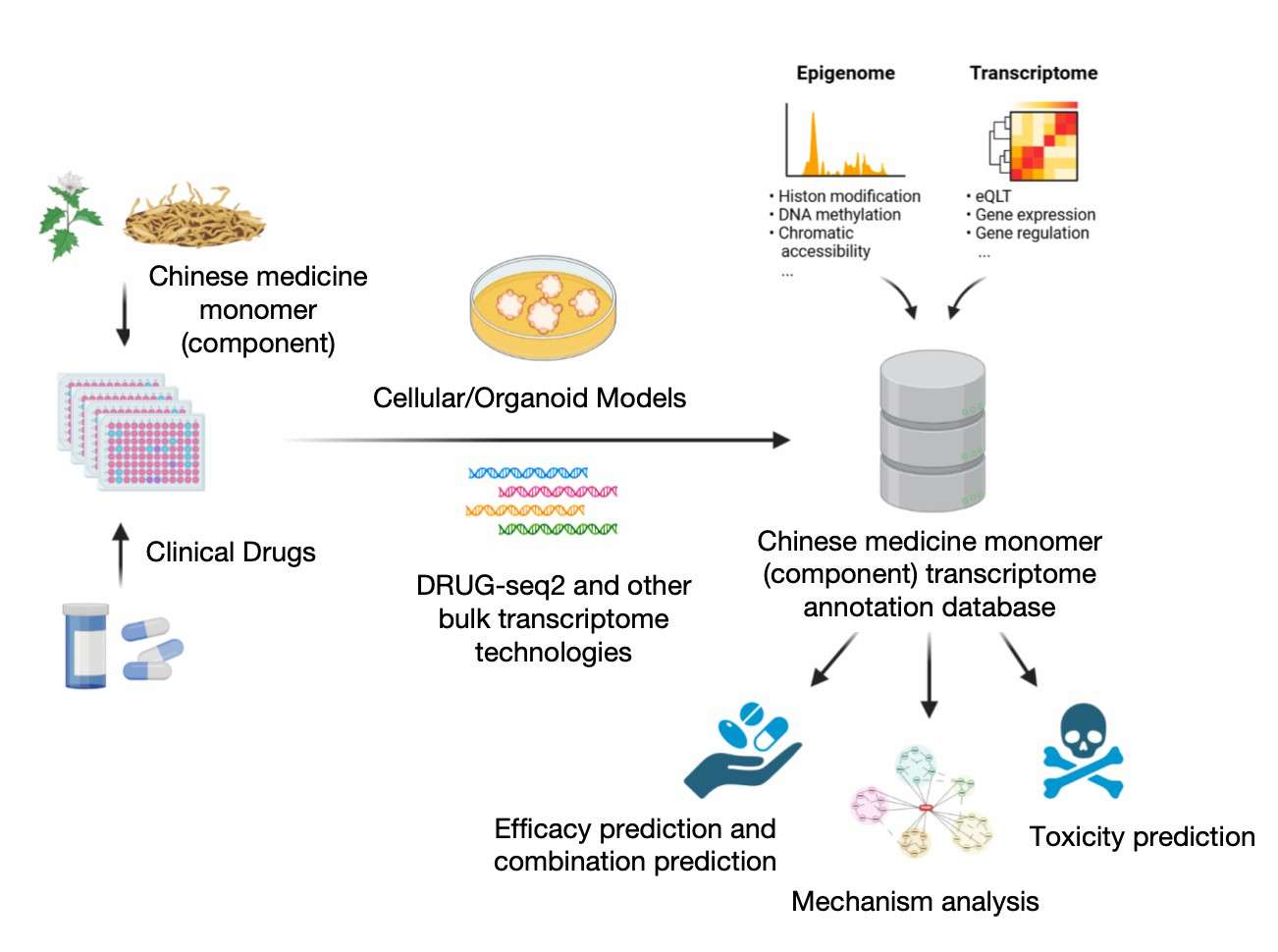 Pharmacodynamic, mechanistic and toxicity studies of traditional Chinese medicine using DRUG-seq2
Pharmacodynamic, mechanistic and toxicity studies of traditional Chinese medicine using DRUG-seq2
Furthermore, DRUG-seq2 is instrumental in the safety evaluation of TCM. Through the analysis of the transcriptome of TCM components, researchers can identify potential toxic responses at an early stage. This is imperative for toxicity prediction of TCM components and can avert potential safety hazards during drug development. Utilising this technique facilitates more precise verification of the pharmacological effects of traditional Chinese medicines, providing a substantial foundation for the modernisation and transformation of Chinese medicine components.
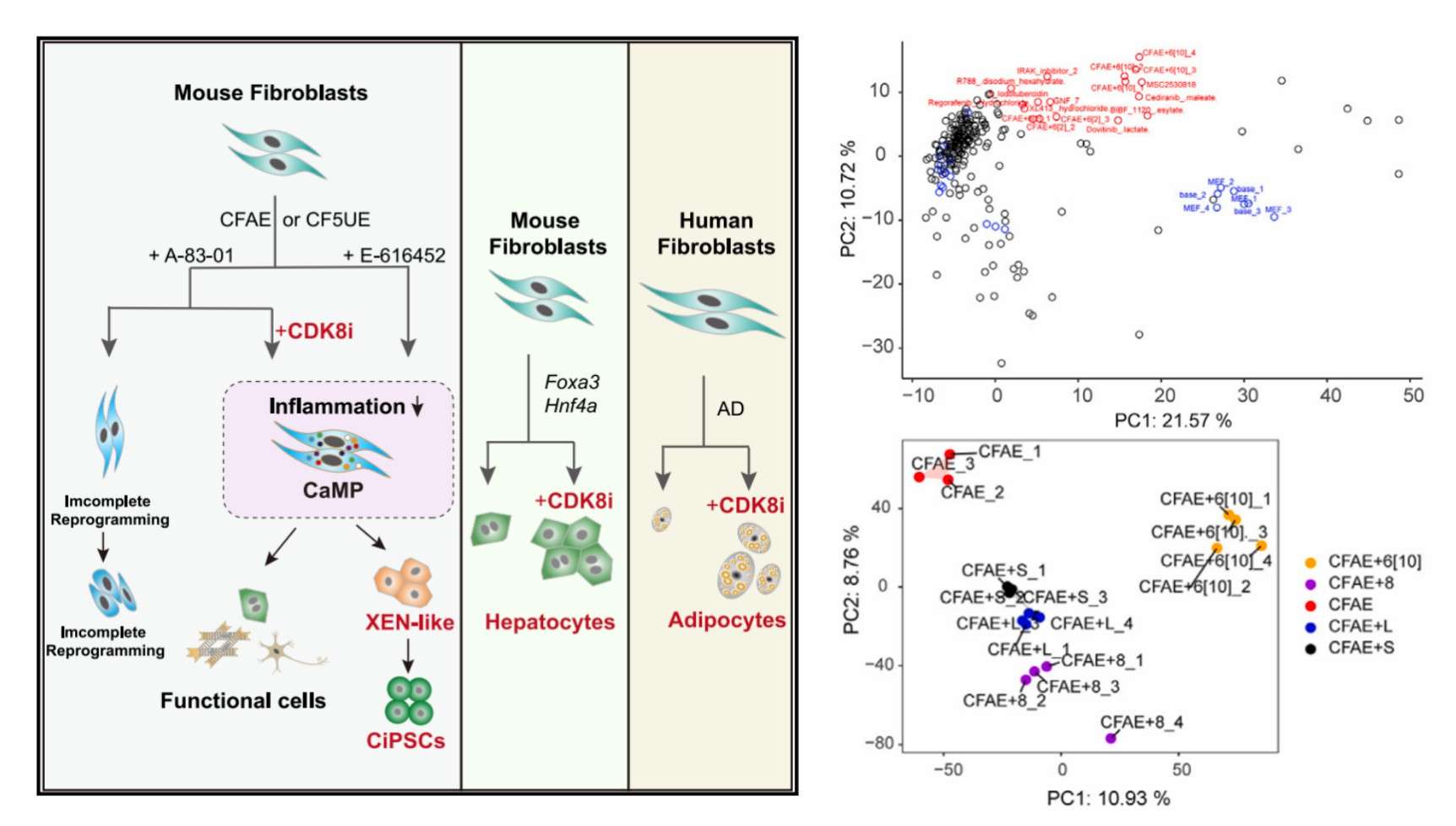 DRUG-seq2-based screening identifies potential drug targets (Li et al., 2023)
DRUG-seq2-based screening identifies potential drug targets (Li et al., 2023)
Finally, significant progress has been made in the application of DRUG-seq2 in the field of drug target discovery. Through the process of compound screening and the analysis of transcriptome clustering, DRUG-seq2 was able to identify key small molecules and drug targets. For instance, researchers screened with this technology to discover the small molecule E-616452, which initiates cellular reprogramming by targeting CDK8. This finding not only offers novel concepts for drug target screening but also further substantiates the pivotal role of DRUG-seq2 in precision drug development.In summary, DRUG-seq2 technology offers a comprehensive and precise instrument for drug discovery, drug target discovery and traditional Chinese medicine research, thereby enhancing the efficiency and safety of new drug discovery and development and possessing a broad spectrum of potential applications.
DRUG-seq2 technology's batch cellular transcriptome assay facilitates the comprehensive comparison of the effects of diverse cell culture components or their combinations, at a minimal cost and with optimal efficiency. This technology platform facilitates a systematic assessment of the effects of diverse culture conditions on cell growth, differentiation and function. This is achieved by a comprehensive analysis of gene expression data from a large number of cell samples, evaluated across a range of dimensions, including the extracellular matrix, signalling factors, small molecule inhibitors, metabolites and nutrients.The massively parallel testing capability of this technology not only optimises the formulation of culture solutions, but also reveals the molecular characteristics of cells in different physiological or pathological states under specific culture conditions. To illustrate this point, consider the following example: by comparing culture media containing different combinations of small molecule inhibitors or metabolites, researchers can discover which components best promote cell differentiation, maintain stem cell status or induce organoid formation.
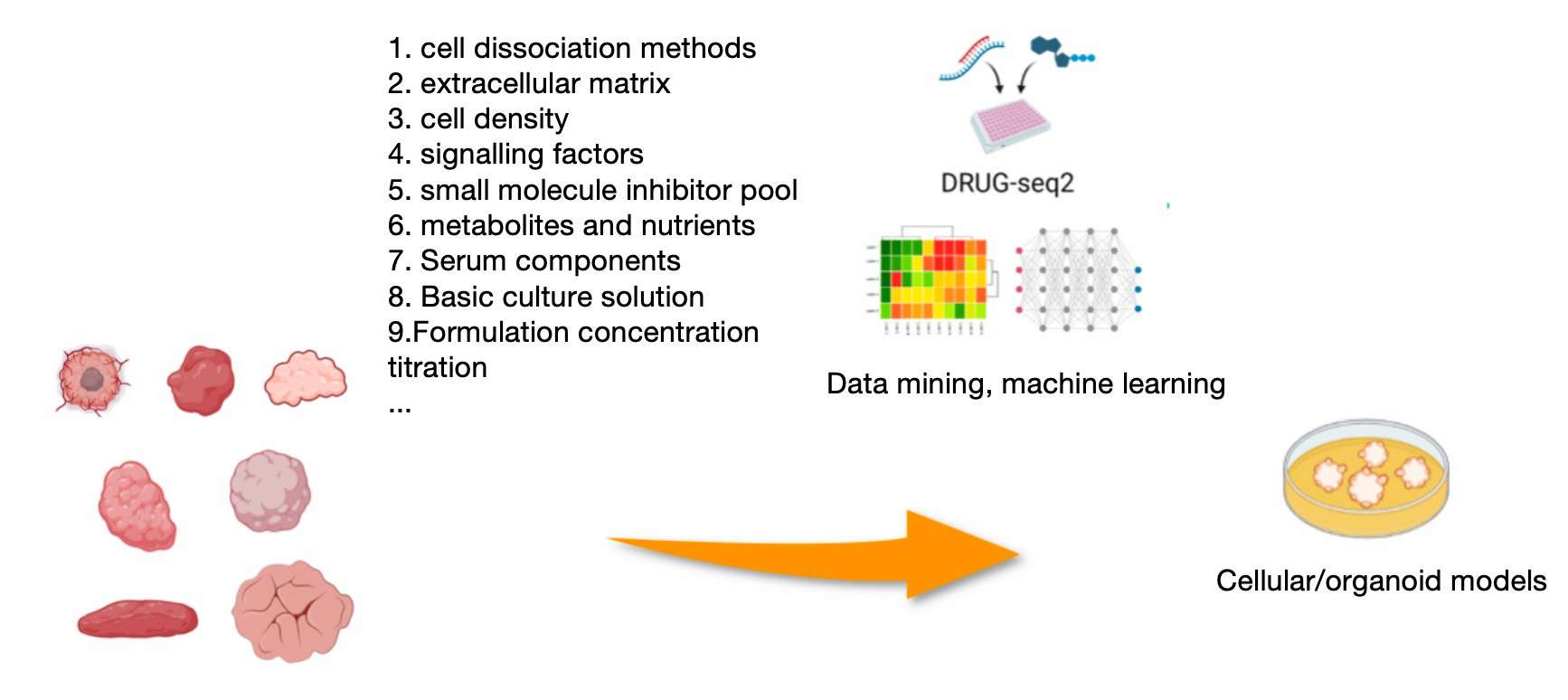 Development and optimisation of culture medium formulations for specific cell populations and organoids using DRUG-seq2
Development and optimisation of culture medium formulations for specific cell populations and organoids using DRUG-seq2
The data measured by DRUG-seq2 can also be used to find the complex correlations hidden between culture conditions and cell transcriptome changes through big data mining and machine learning. Such data-driven analyses can help researchers determine the optimal combination of culture conditions so that cell or organoid models can better reflect human physiology or disease characteristics, thus enhancing the value of the models in drug screening, disease mechanism research and regenerative medicine. The development of these optimised models will facilitate the provision of more realistic and clinically relevant tools for scientific research and drug development, thereby accelerating the transition from laboratory to clinical applications.
References

CD Genomics is transforming biomedical potential into precision insights through seamless sequencing and advanced bioinformatics.
We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy