We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy

Gene editing technology, which involves the modification of target genes within the genome through deletion or addition, has revolutionized the field of biomedical science. Among these technologies, the CRISPR/Cas9 system has emerged as a superior tool compared to traditional gene editing methods such as Zinc Finger Nucleases (ZFNs) and Transcription Activator-Like Effector Nucleases (TALENs). CRISPR/Cas9 offers advantages in terms of simplicity, cost-effectiveness, and higher editing efficiency.
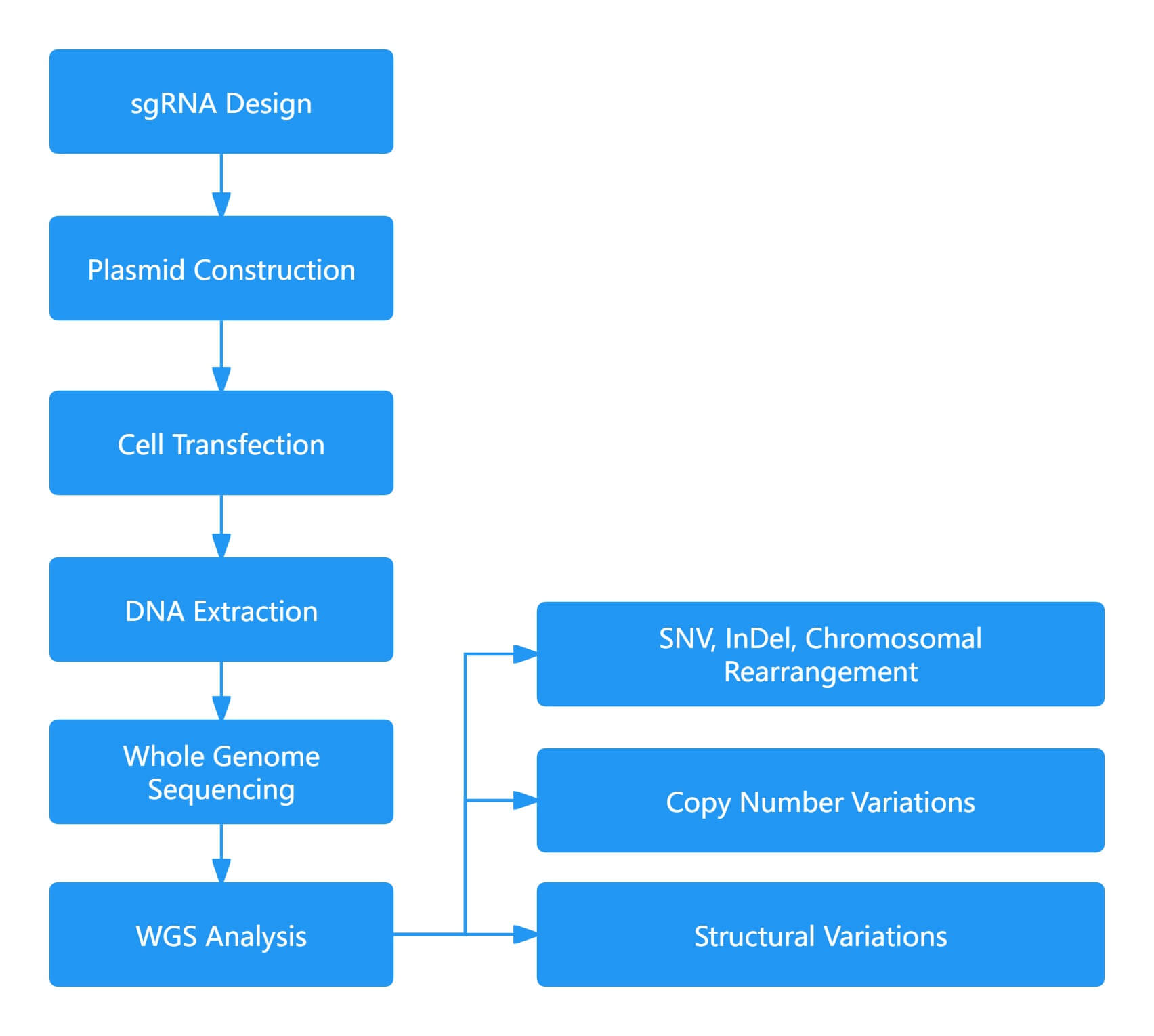 Figure 1. Major concerns/outcomes of off‐target effects
Figure 1. Major concerns/outcomes of off‐target effects
In vitro detection methods are essential for evaluating the off-target effects and specificity of CRISPR/Cas9 and other programmable nucleases outside of a living organism. These methods provide a controlled environment to assess how these nucleases interact with DNA, allowing for a detailed analysis of their cutting efficiency and potential off-target activities. Below are several key in vitro detection methods:
WGS is a comprehensive and unbiased method for evaluating mutations. However, it is only capable of detecting a small fraction of off-target cells within transgenic cell populations and cannot distinguish between single nucleotide variants (SNVs) resulting from sequencing errors or naturally occurring mutations. Therefore, it is crucial to thoroughly investigate the genetic background before assessing off-target effects. WGS has been employed to detect off-target mutations induced by Cas9 in various plant species, including Arabidopsis thaliana, rice, tomato, and cotton. Nevertheless, WGS is highly reliable only for detecting off-target sites with higher frequencies and lacks the sensitivity required for detecting off-target sites in large populations. When performing WGS on genomic DNA from many independent cells, it has the potential to detect all types of off-target base editing across cells or entire higher organisms.
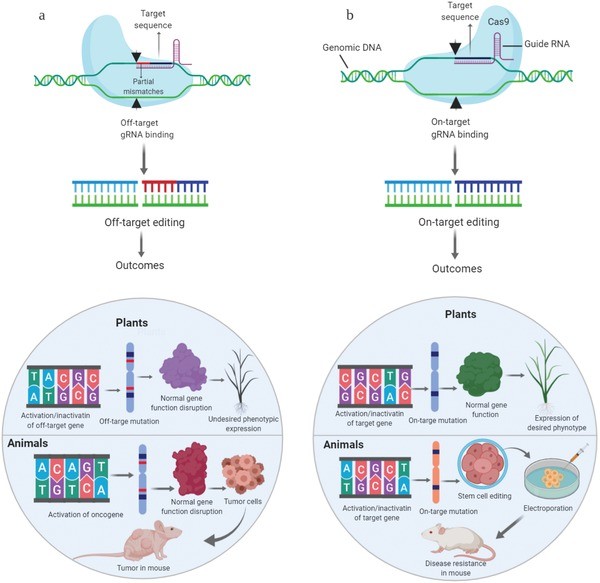 Figure 2. Whole Genome Sequencing for Off-Target Analysis
Figure 2. Whole Genome Sequencing for Off-Target Analysis
Digenome-seq represents an unbiased, robust, economical, sensitive, and reproducible approach for analyzing genome-wide off-target effects induced by Cas9 and other programmable nucleases. Unlike methods that rely on DNA binding, Digenome-seq is based on DNA cleavage, making it suitable for application across the entire genomic landscape. Notably, Digenome-seq captures potential off-target sites through RNA/DNA bulge capture, enhancing its sensitivity. This method is sufficiently sensitive to detect off-target levels even when induced indexes occur at a frequency as low as 0.1%, which is near the detection limit of high-throughput sequencing strategies.
For a detailed exploration, please refer to the technical review: Analysis of Genome-Wide CRISPR-Cas9 Off-Target in Human Cells Using Digenome-seq.
SITE-seq (Selective Integration of Targeted Endonucleases Sequencing) is a biochemical approach designed to identify Cas9 cleavage sites within purified genomic DNA. This method relies on sequencing tagged genomic DNA ends and selective enrichment to pinpoint these cleavage sites. In the SITE-seq protocol, the genomic DNA undergoes digestion with varying concentrations of single-guide RNA-protein complexes (sgRNPs), allowing the recovery of off-target sites with both low and high cutting sensitivities. The method also involves the development of sequencing libraries highly enriched for sgRNP cleavage fragments, enabling specific analysis with minimal read depth. This capability is crucial for employing SITE-seq as a high-throughput tool for guide RNA selection.
PEM-seq (Primer-Extension-Mediated Sequencing) is employed to characterize off-target sites and other chromosomal abnormalities, such as genome-wide translocations, small deletions, and large deletions induced by Cas9. PEM-seq has also been utilized to evaluate various methods aimed at reducing Cas9 off-target activity, providing a comprehensive assessment of CRISPR/Cas9 specificity and editing efficiency.
For a detailed review, refer to: PEM-seq: Optimizing Gene Editing and Tracking DNA Repair Methods
In-cell detection methods are employed to assess the activity of nucleases, such as CRISPR/Cas9, within a cellular environment. These methods allow for the evaluation of off-target effects and editing efficiency directly in living cells, providing a more accurate representation of nuclease behavior in physiological conditions. The following sections describe key in-cell detection methods used in CRISPR research:
DISCOVER-Seq is a powerful and sensitive method designed for unbiased identification of off-target sites in cellular models. This approach has been employed in vivo, such as in studies involving adenoviral gene editing in mouse liver, paving the way for real-time detection of off-target effects during therapeutic gene editing. DISCOVER-Seq has been utilized with various types of Cas nucleases and reliably detects off-target editing events across multiple gRNAs and Cas nucleases in both human and mouse cells. It is also applicable to other species with large genomes.
For further details, please refer to: DISCOVER-Seq In Vivo Unbiased Detection of CRISPR Off-Targets Using DISCOVER-Seq
EndoV-Seq is a widely used method for comprehensive research on ABE (Adenine Base Editors) specificity across the genome. This technique involves the deamination of genomic DNA by EndoV (Endonuclease V) followed by WGS. EndoV-Seq is employed to evaluate the targeting and off-target effects of ABEs and has provided insights into enhancing ABE specificity. The process utilizes EndoV, derived from Thermotoga maritima, to cleave DNA strands containing inosine, a deaminated product of adenine. The processed samples are then subjected to WGS to identify off-target sites.
CD Genomics has established an efficient and mature platform for genome editing, high-throughput sequencing, and bioinformatics analysis. We offer comprehensive and precise CRISPR/Cas9 off-target detection and analysis services.
Reference

CD Genomics is transforming biomedical potential into precision insights through seamless sequencing and advanced bioinformatics.
We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy