The introduction to non-coding RNAs
Non-coding RNAs (ncRNAs) used to be considered as transcription noises or byproducts of RNA processing, but increasing evidence suggests that a majority of them are biologically functional and regulate various activities in the cells. The ncRNAs are roughly classified into two categories according to their sequence length: small ncRNAs (<200 bp) and long ncRNAs (200 bp or more). The categories of ncRNA are listed in Table 1.
Table 1. Overview of ncRNA (Fu 2014).
| ncRNAs |
Full name |
Function |
| Housekeeping ncRNAs |
| rRNA |
Ribosomal RNA |
Translational machinery |
| tRNA |
Transfer RNA |
Amino acid carriers |
| snRNA |
Small nuclear RNA |
RNA processing |
| snoRNA |
Small nucleolar RNA |
RNA modifications |
| TR |
Telomere RNA |
Chromosome end synthesis |
| Regulatory ncRNAs |
| miRNA |
MicroRNAs |
RNA stability and translation control |
| endo-siRNA |
Endogenous siRNA |
RNA degradation |
| rasiRNA |
Repeat-derived RNA |
Transcriptional control |
| piRNA |
Piwi-interacting RNA |
Silencing transposon and mRNA decay |
| eRNA |
Enhancer-derived RNA |
Regulation of gene expression |
| PATs |
Promoter-associated RNA |
Transcription initiation and pause release |
| lncRNA |
Long non-coding RNA |
Imprinting, epigenetics, nuclear structure |
As shown in Table 1, ncRNAs can be roughly divided into two classes: housekeeping ncRNAs and regulatory ncRNAs. Housekeeping ncRNAs, involving rRNA, tRNA, snRNA, snoRNA, and TR, are considered “constitutive” since they are ubiquitously expressed in all cell types and offer essential functions to the organisms. Regulatory ncRNAs, involving miRNA, endo-siRNA, rasiRNA, piRNA, eRNA, PATs, and lncRNA, have received increasing attention from the research community due to their regulatory function in gene expression, imprinting, and epigenetics. RNA-seq is an advanced technique to illustrate the ncRNA species. Here, we made a summary of the bioinformatics tools for ncRNA analysis with data from NGS.
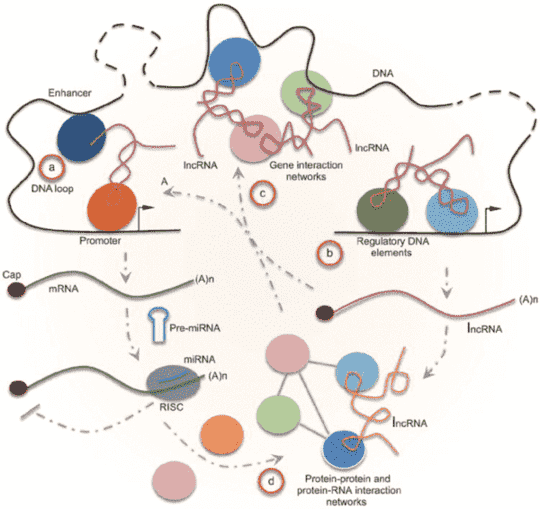
Figure 1. ncRNAs as integrated parts of gene network (Fu 2014).
Small ncRNA analysis
Small RNAs play a crucial role in transcriptional regulation and are essential to fully understand the entire scenario of transcriptional regulation. Their aberrant expression profiles are considered to be associated with cellular dysfunction and disease. Therefore, many researches are focused on detection, prediction, or expression quantification of small RNAs, particularly miRNAs, to better understand human health and disease. The available computational tools for small RNA sequencing data are summarized in Table 2.
Table 2. Computational tools for small ncRNA analysis
| Tools |
Descriptions |
| DARIO |
Quantify and annotate ncRNAs with access to several ncRNA public databases. |
| CPSS |
Quantify and annotate ncRNAs, with special emphasis on miRNAs. |
| ncPRO-seq |
Detect known small ncRNAs in an unbiased way and discover novel ncRNA species. |
| CoRAL |
Divide small ncRNA into functional categories based on biologically interpretable features other than sequence;
Annotate ncRNA in less well-characterized organisms. |
| RNA-CODE |
Combine secondary structure with de novo assembly.
Applicable to ncRNA annotation lacking reference genomes. |
| miRDeep |
Used to detect both known and novel miRNAs in small RNA sequencing data. |
Circular RNA detection
CircRNAs are a novel type of RNA that form a covalently closed continuous loop. Most of them are generated from exonic or intronic sequences, and RNA-binding proteins (RBPs) or reverse complementary sequences are necessary for their biogenesis. CircRNAs are mostly conserved, and function as miRNA sponges, regulator of splicing and transcription, or modifiers of parental gene expression. Increasing evidence suggests the potential significance of circRNA in human diseases, such as atherosclerotic vascular disease, neurological disorders, and cancer. Among all the presented tools for circRNA detection, CIRI, CIRCexplorer, and KNIFE exhibit a balanced performance between precision and sensitivity. The available computational tools for circRNA sequencing data are summarized in Table 2.
Table 3. Computational tools for circular RNA detection.
| Method |
Approach |
dependencies |
| CIRI |
Segmented read-based |
Bwa, peri |
| CIRCexplorer |
Segmented read-based |
STAR, bedtools, python (pysam, docopt, Interval) |
| KNIFE |
Candidate-based |
Bowtie, Bowtie2, tophat2, samtools, perl |
LncRNA investigation
LncRNA is a type of non-coding RNA with more than 200 nucleotides, such as lincRNAs and macroRNAs. LncRNAs function as a platform for the interaction with mRNA, miRNA, or protein. They have emerged as vital regulators in diverse aspects of biology, including transcriptional regulation, post-transcriptional regulation, and chromatin remodeling. Increasing researches suggest misexpression of lncRNAs contributes to tumor initiation, growth, and metastasis. LncRNAs hence become a promising target for cancer diagnosis and therapy. The combination of lncRNA sequencing and matched computational tools is a powerful approach for this purpose.
Table 4. Computational tools for lncRNA investigation.
| Tools |
Applications |
Reference |
| lncRScan |
Detect lncRNA from the complex assemblies; Distinguish lncRNA from mRNAs |
(Sun et al., 2012) |
| iSeeRNA |
Accurately and quickly detect lincRNA from large datasets |
(Sun et al., 2013) |
| Annocript |
Detect lncRNA by leveraging public databases and sequence analysis software to verify high non-coding potential |
(Musacchia et al. 2015) |
| LncRNA2Function |
Annotate lncRNA based on the theory that similar expression patterns across diverse conditions may share similar functions and biological pathways. |
(Jiang et al. 2015) |
References:
- Choudhuri S. Small noncoding RNAs: biogenesis, function, and emerging significance in toxicology. Journal of biochemical and molecular toxicology, 2010, 24(3): 195-216.
- Fu X D. Non-coding RNA: a new frontier in regulatory biology. National science review, 2014, 1(2): 190-204.
- Gao Y, Wang J, Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome biology, 2015, 16(1): 4.
- Jiang Q, Ma R, Wang J, et al. LncRNA2Function: a comprehensive resource for functional investigation of human lncRNAs based on RNA-seq data//BMC genomics. BioMed Central, 2015, 16(3): S2.
- Musacchia F, Basu S, Petrosino G, et al. Annocript: a flexible pipeline for the annotation of transcriptomes able to identify putative long noncoding RNAs. Bioinformatics, 2015, 31(13): 2199-2201.
- Qu S, Yang X, Li X, et al. Circular RNA: a new star of noncoding RNAs. Cancer letters, 2015, 365(2): 141-148.
- Su Y, Wu H, Pavlosky A, et al. Regulatory non-coding RNA: new instruments in the orchestration of cell death[J]. Cell death & disease, 2016, 7(8): e2333.
- Sun K, Chen X, Jiang P, et al. iSeeRNA: identification of long intergenic non-coding RNA transcripts from transcriptome sequencing data. BMC genomics, 2013, 14(2): S7.
- Sun L, Zhang Z, Bailey T L, et al. Prediction of novel long non-coding RNAs based on RNA-Seq data of mouse Klf1 knockout study. BMC bioinformatics, 2012, 13(1): 331.
- Veneziano D, Nigita G, Ferro A. Computational approaches for the analysis of ncRNA through deep sequencing techniques. Frontiers in bioengineering and biotechnology, 2015, 3: 77.
- Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms, 2014, 1839(11): 1097-1109.
- Zeng X, Lin W, Guo M, et al. A comprehensive overview and evaluation of circular RNA detection tools. PLoS computational biology, 2017, 13(6): e1005420.
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.


 Sample Submission Guidelines
Sample Submission Guidelines

