Genomics is driving rapid advances in the field of cancer research, particularly in the way cancer is diagnosed, treated and monitored. Whole exome sequencing (WES) is a genetic analysis method that uses sequence capture technology to capture and enrich DNA from whole exome regions for high-throughput sequencing. Exome sequencing is simpler, more cost-effective and more efficient than whole genome sequencing, and its target region coverage is higher, facilitating variant detection.
Mutations in whole-exome regions of the cancer genome are most likely to affect tumor development. Accurate detection of SNVs and InDels by WES can efficiently resolve cancer susceptibility and pathogenicity genes, providing a reliable method for cancer genomic research. Whole-exome sequencing has become the method of choice for many tumor-normal sample comparisons.
With the increasing depth and understanding of the tumor exome, there are many other points of analysis besides driver gene and mutation characterization he tumor heterogeneity analysis of exome sequencing results. The following are some cases.
Tumor mutational burden (TMB) analysis using WES
Tumor Mutational Burden (TMB), defined as the number of somatic mutations per MB of the longest transcript sequence. The current state of research is that although TMB is a potential biomarker for immunotherapy of solid tumors, there is still a lack of consensus on the optimal TMB threshold for specific solid tumors, and a more refined differentiation of the optimal TMB threshold could greatly improve the therapeutic tools for certain refractory tumors. By analyzing the relationship between WES and TMB for all 32 cancer types combined and alone, researchers have found that certain cancer types (e.g., uterine, bladder, and colon cancers) exhibit greater variability in panel TMB values compared to lung, and head cancers.
Homologous recombination defect (HRD) analysis using WES
Homologous recombination (HR) is a highly conserved process that plays an important role in DNA repair, DNA replication, meiotic chromosome segregation, and telomere maintenance. In general, HRD can be caused by many factors such as germline mutations or somatic mutations in HRR-related genes and epigenetic inactivation. For example, somatic mutations in BRAC1 and BRCA2, which encode homologous recombinant proteins, have been popularly studied in HRD because of their involvement in hereditary breast and ovarian cancers.
By detecting the HRD status of cancer cells through exon sequencing, HRD can be used as a new biomarker to play a great role in the individualized treatment of tumors. Due to the sensitivity of HRD cells to PARP inhibitors, the detection of HRD is crucial in the drug guidance and prognosis monitoring of tumor patients. The therapeutic effect of HRD as a tumor marker for colorectal cancer (CRC) was investigated by WES analysis of 9321 patients with CRC tumors. It was found that 1270 (13.6%) cases were HRD positive and the rest were HRD-free (i.e., HRP). the incidence of HRD tumors was higher in MSI tumors than in MSS tumors. In the TRIBE2 study, overall survival was longer in patients with MSS and HRD tumors than in patients with MSS and HRP tumors. Therefore, HRD is a specific marker of MSS CRC tumors with specific molecular and prognostic features.
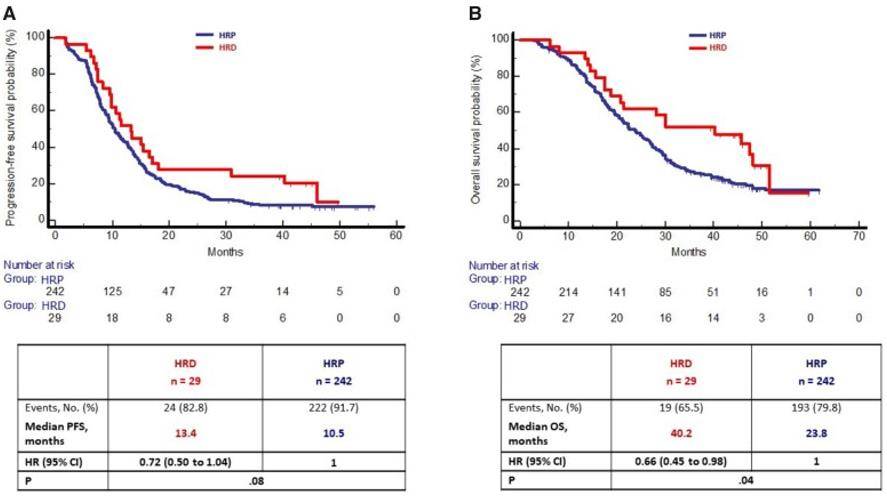 Homologous recombination deficiency alterations in colorectal cancer (Moretto R et al. 2022)
Homologous recombination deficiency alterations in colorectal cancer (Moretto R et al. 2022)
Microsatellite instability analysis (MSI) using WES
Microsatellite (MS) are short tandem repeats (STR) or simple sequence repeats (SSR) consisting of 1-6 bp repeating base units, uniformly distributed in eukaryotic genomes, including coding and non-coding regions. Numerous studies have shown that MSI is closely associated with tumorigenesis, and it has been clinically used as an important molecular marker for prognosis and development of adjuvant treatment plans for solid tumors such as colorectal cancer.
In MSI analysis, we can analyze the MS status of a single sample; we can also screen for high-frequency MSI genes and predict the microsatellite status of tumor samples based on the analysis of microsatellite loci at the somatic cell level.
The authors studied the MSI status of 96 constitutional mismatch repair deficiency (CMMRD) cancers and 8 normal tissues. It was found that only 17 of the 96 CMMRD tumors had a high MSI status (MSI-H), a result that was independent of the specific MMR mutation gene, and that different tumors in the same patient resulted in different MSI classifications, while correlating with tissue specificity, such as the highest percentage of MSI-H in gastrointestinal cancers compared to brain tumors.
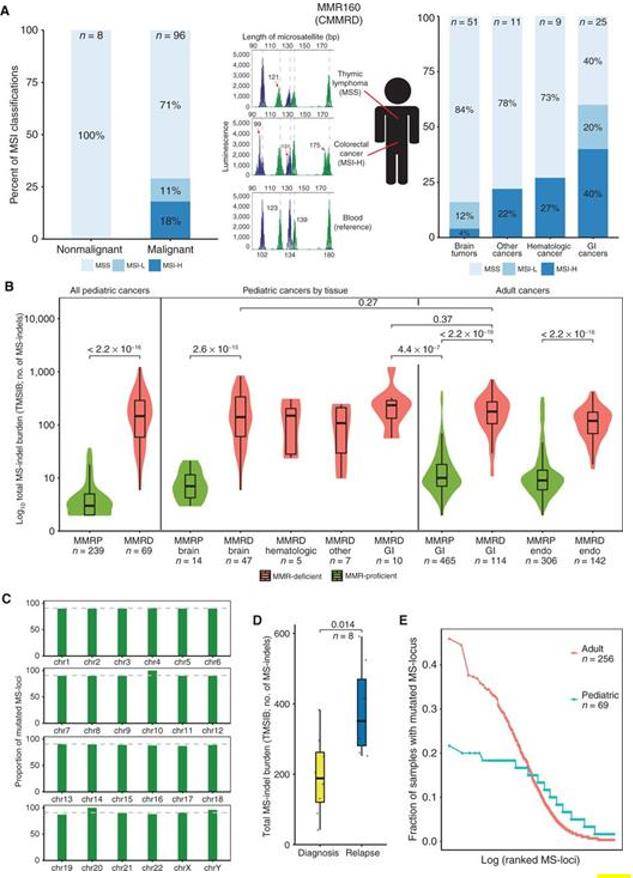 MSs accumulate MS-indels in germline MMRD cancers (Chung J et al. 2021)
MSs accumulate MS-indels in germline MMRD cancers (Chung J et al. 2021)
Summary
The application of whole exome sequencing in tumor research can provide ideas for diagnosis, treatment, monitoring, individualized treatment through tumor biomarkers, and also provide research methods to study the mechanism of action of therapeutic tumor drugs. Based on the advantages of low cost and high efficiency, WES has a considerable application background in the study of tumor mechanism, and looking into the future, WES will become an effective tool for molecular biology and individualized treatment of tumors.
References:
- Merino D M, McShane L M, Fabrizio D, et al. Establishing guidelines to harmonize tumor mutational burden (TMB): in silico assessment of variation in TMB quantification across diagnostic platforms: phase I of the Friends of Cancer Research TMB Harmonization Project. Journal for immunotherapy of cancer, 2020, 8(1).
- Moretto R, Elliott A, Zhang J, et al. Homologous recombination deficiency alterations in colorectal cancer: Clinical, molecular, and prognostic implications. JNCI: Journal of the National Cancer Institute, 2022, 114(2): 271-279.
- Chung J, Maruvka Y E, Sudhaman S, et al. DNA Polymerase and Mismatch Repair Exert Distinct Microsatellite Instability Signatures in Normal and Malignant Human Cells. Cancer discovery, 2021, 11(5): 1176-1191.
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.


 Sample Submission Guidelines
Sample Submission Guidelines
 Homologous recombination deficiency alterations in colorectal cancer (Moretto R et al. 2022)
Homologous recombination deficiency alterations in colorectal cancer (Moretto R et al. 2022) MSs accumulate MS-indels in germline MMRD cancers (Chung J et al. 2021)
MSs accumulate MS-indels in germline MMRD cancers (Chung J et al. 2021)