Breeding Tools: Common Database for Rice Research
Rice (Oryza) sativa is the main food source for half of the world's population, and contributes about 20% calories to human diet. Under the background of rapid global population growth and climate change, rice breeders are faced with the challenge of developing new sustainable rice varieties. These varieties not only need to increase yield, but also pay attention to the nutritional quality of grains and reduce environmental impact. Since the international rice genome sequencing project released the first "gold standard" reference genome of Japanese fine varieties in 2005, with the advancement of global research, more and more rice material genomes have been successfully sequenced, assembled and annotated, and rice pan-genome research has gradually increased. The accumulation of these genomic resources has greatly promoted the development of rice genetics and breeding research, and provided key data support for cultivating better quality and adaptability rice varieties. In order to better support rice research, there collates the database resources commonly used in rice research, hoping that these valuable tools can provide strong support for scientific research.
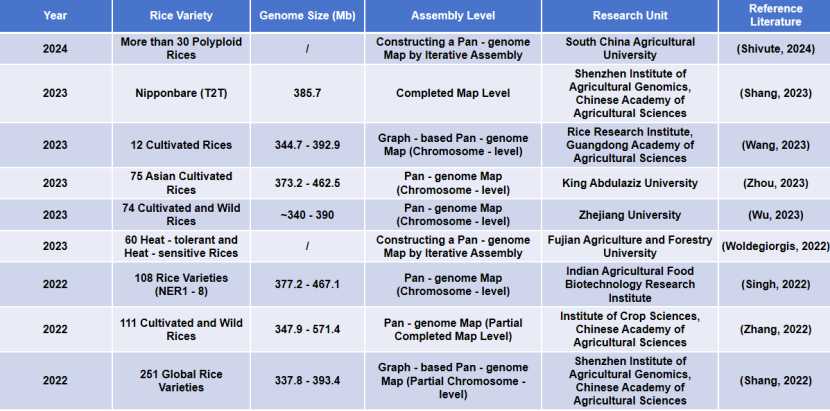 Research progress of rice genome
Research progress of rice genome
Service you may interested in
Comprehensive Rice Research Databases
RAP-DB Database
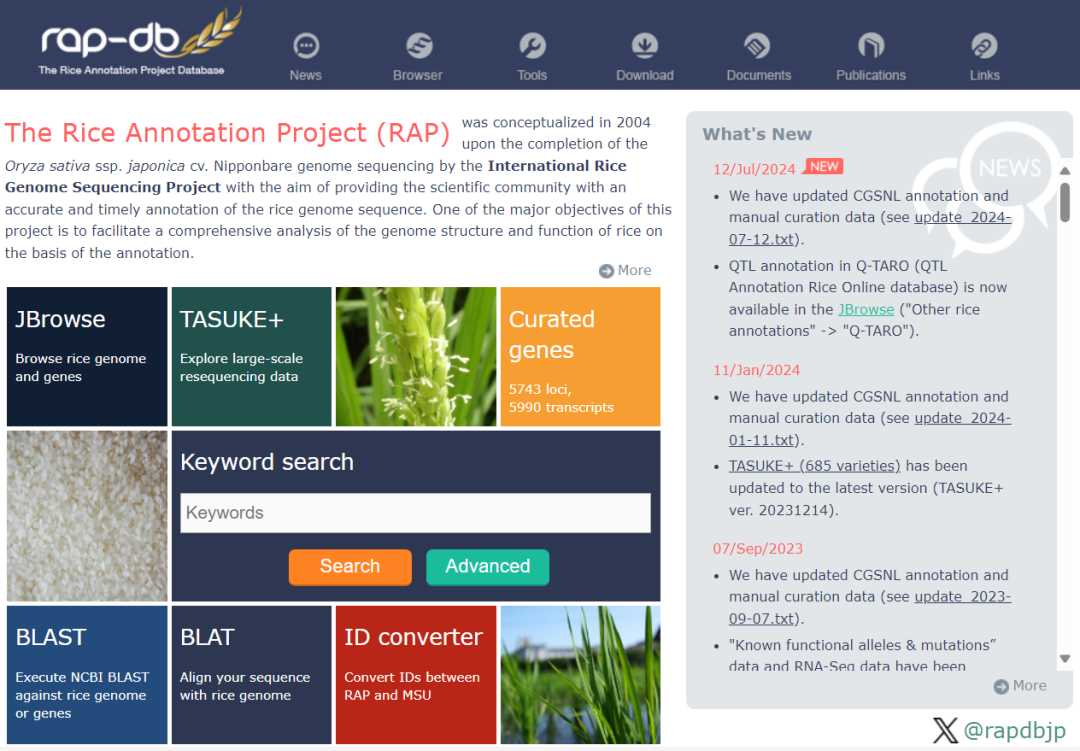
The rice annotation project database RAP-DB (https://rapdb.dna.affrc.go.jp/) was launched in 2004, aiming at providing accurate annotation of rice genome and promoting in-depth analysis of its structure and function. In addition, users can use GBrowse to view the gene distribution of Nissin IRGSP-1.0, search the gene function and sequence by gene name, use BLAST to search for homologous genes, and use ID Converter to convert gene ID.
RGAP Database
The rice genome annotation project database RGAP (https://rice.uga.edu/) of Michigan State University mainly provides rice genome sequence and annotation data. The database takes Nissin as a reference genome, including genome data and CDS data. Users can convert gene names through gene ID and RAP-db, and carry out BLAST search using known sequences to obtain information such as loci number, gene structure and CDS sequence of genes. On September 3, 2024, the database was updated, the genome browser was upgraded to Jbrowse2, and the gene expression and co-expression data and web pages were also updated. In addition, the website has added a syntelogs page, which contains two new syntelog data sets.
Gramene Database
Gramene (https://www.gramene.org/), a comparative genomics database, integrates various data resources through comparative functional genomics, and lists the genome information of different rice subspecies, including Oryza rufipogon, Oryza nivara and Oryza glaberrima. Plant provided by this website Reactome connects the reported gene signal networks in series to form a signal network about growth and development signals, secondary metabolic pathways and biotic or abiotic stresses. Users can view specific data on the website.
RPAN Database
The rice pan-genome database RPAN (https://cgm.sjtu.edu.cn/3kricedb/index.php) is constructed from 3K rice genome, and contains about 370Mbp of IRGSP genome and about 260Mbp of new sequences. The database provides basic information of 3,010 rice germplasm, including the sequence and gene annotation of rice pan-genome, gene presence-deletion variation (PAV) and expression profile. Its basic search function allows users to query the basic information and sequencing prospect of a single gene or rice. The advanced search function supports searching the shared genes of multiple rice germplasm or the existence of multiple genes. Visualization functions include tree browser for viewing the phylogeny of 3K rice germplasm and genome browser for gene annotation and PAV.
MBKbase Database
MBKbase (https://www.mbkbase.org/rice), a comprehensive database of rice molecular breeding, provides the genotype and related phenotype of each gene locus in the population by integrating the high-quality reference genome of rice, reveals the relationship among germplasm, phenotype and genotype, and realizes online integration analysis and visualization of genotype and phenotype. The database consists of five functional modules: the germplasm module provides information on rice germplasm resources, the phenotype module displays trait records, the genotype module integrates sequenced samples and SNP/InDel information to confirm the verified alleles, and the population module provides information on WGS population and its phylogenetic tree, which provides important support for functional genomics research and molecular breeding of rice.
SNP-Seek Database
Rice SNP retrieval database SNP-Seek (https://snpseek.irri.org/index.zul) was established by the International Rice Research Institute in 2014, which mainly provides information on rice genotypes, phenotypes and varieties. The database is based on the Japanese reference genome IRGSP-1.0, integrates SNP genotyping data from 3,000 rice genome projects, and includes phenotypic data from the International Rice Gene Bank Collection Information System (IRGCIS). Its purpose is to centralize the information access of rice research data and provide calculation tools to discover new gene-trait associations and promote rice improvement.
Oryzabase Database
Oryzabase (https://shigen.nig.ac.jp/rice/oryzabase/), a comprehensive rice science database, was established by the Japan Rice Research Council in 2000 to collect as much information about rice as possible, including strain inventory, mutant information, chromosome map, gene dictionary and basic knowledge. The database covers the research progress from classical rice genetics to the latest genomics, provides a wide range of genetics and genomics data, involves many popular research topics, and provides rich information for rice researchers.
Rice Gene-related Databases
RGI Database
Rice Gene Index Database RGI (https://riceome.hzau.edu.cn/) contains reference genomes of 16 major Asian rice materials, and processes multiple genomes and annotation information in a unified way. A total of 119,783 non-redundant genomes were identified to represent the whole Asian rice gene set, and a unified digital orthogonal gene index (OGI) was also established, indicating its representativeness in all materials. RGI provides a wealth of modules and tools to help researchers query and visualize rice genes and their homologous relationships. Each gene has a "comprehensive graphic information card", which contains homologous gene index, sequence and function information, and shows transcript structure and phylogenetic tree.
RiceVarMap v2.0 Database
Rice Genome Variation and Functional Annotations Comprehensive Database RiceVarMap v2.0 (http://ricevarmap.ncpgr.cn/) provides information of 17,397,026 genome variations, including 14,541,446 SNPs and 2,855,580 small INDEL, from 4,726 sequencing data of rice materials. These mutations were identified by GATK software based on OS-nipponbare-reference-irgsp-1.0, ensuring high quality and complete genotype data. In addition, the database also contains phenotypic data and GWAS results to help researchers search for important SNPs related to various traits. The new version introduces more powerful query and visualization tools, such as online forecasting tool Regulatory Variant.
RiceXPRo Database
RiceXPRo (https://ricexpro.dna.affrc.go.jp/) is a repository of rice gene expression profiles, covering the whole growth process of rice plants in natural field conditions, rice seedlings treated with different plant hormones, and specific cell types and tissues separated by laser microdissection (LMD). The database aims to characterize the expression profiles of all predicted genes in rice and provide reference information for functional genomics. All expression profiles were generated using a single microarray platform, and the probes were based on the manually arranged gene model in RAP-DB and the full-length cDNA sequence information of rice in KOME database.

RiceENCODE Database
RiceENCODE (http://glab.hzau.edu.cn/RiceENCODE/index.html) is a comprehensive encyclopedia database of rice DNA elements, which combines published three-dimensional interactive data (ChIA-PET, Hi-C) and epigenome data sets (ChIP-Seq, ATAC-Seq, MNase-Seq, FAIRE-Seq, WGBS, RNA-Seq). The database contains 694 data sets, which is the largest rice epigenome data at present.
RiceGE Database
Rice Functional Genome Expression Database RiceGE (http://signal.salk.edu/cgi-bin/RiceGE5) focuses on providing rice-related gene expression patterns and gene function annotation information. The database integrates a large number of transcriptome data generated based on high-throughput sequencing technology, and supports users to query the expression of specific genes at different developmental stages or in response to environmental stimuli. In addition, it also contains multi-dimensional information such as gene cloning status and gene function verification, which provides valuable resources for researchers to deeply understand the function and regulation mechanism of rice genes
RiceRelativesGD Database
Rice-related species genome database RiceRelativesGD (http://ibi.zju.edu.cn/ricerelativesgd/) provides gene and genome resources from 16 rice related species in Gramineae, including important rice weeds such as weedy rice and barnyard grass (Echinochloa crus-galli) and ancient crop Zizania. Latifolia), nine kinds of wild rice and African cultivated rice. The database specifically provides the genes unique to indica and japonica rice in various rice related species and their annotations, and includes online services, such as genome browser, search tool and phylogenetic tree construction tool, to help researchers make better use of the genome data in the database.
funRiceGenes Database
funRiceGenes (http://funricegenes.ncpgr.cn/) is a comprehensive database of rice cloned genes. Through text mining of more than 6,000 published documents, more than 3,000 cloned genes were collected, including more than 400 gene families and more than 200 gene interaction networks. These data provide important clues and references for the study of gene function.
Rice Indica cDNA Database
The indica cDNA database (http://server.ncgr.ac.cn/ricd/index.html) is mainly composed of two parts, among which the full-length indica cDNA database (Guangluai No.4) is developed by the National Gene Research Center of Shanghai Institute of Life Sciences, China Academy of Sciences and maintained by the National Gene Research Center. At present, it contains about 21,960 unique 5'ESTs and 10,081 full-length cDNAs. The estimated full-length cDNA database of indica rice (Minghui 63) was developed by Huazhong Agricultural University, which contains about 27,130 unique ESTs and 12,727 estimated full-length cDNA information. Indica rice and japonica rice are adapted to different climatic regions, and the comparison of their transcriptome will help to understand the molecular mechanism of phenotypic differences and promote the molecular breeding of rice.
RiceFREND Database
Rice Gene Co-expression Database RiceFREND (https://ricefrend.dna.affrc.go.jp/) is a rice gene co-expression database based on a large number of microarray data from different tissues and organs, which covers the data of rice in various growth stages under natural field conditions and includes gene expression information under various plant hormones. These gene expression data for co-expression analysis can be accessed through RiceXPro database, which focuses on gene expression profiles obtained by microarray analysis. RiceFREND aims to provide a platform to identify genes related to functions in various biological pathways and metabolic processes.
Rice Mutant-related and Other Databases
Rice Tos17 Insertion Mutant Database
The Rice Tos17 Insertion Mutant Database (https://tos.nias.affrc.go.jp/), could compare the flanking sequences of rice mutant lines induced by Tos17 by the BLAST search tool, and computer-aided rice mutant screening can be realized. In addition, the database also provides a browsing function, and users can view the specific position of the insertion site in the rice genome and the relevant phenotypic information of each mutant strain.
Oryza Tag Line Database
Rice T-DNA insertion mutation database Oryza Tag Line (https://oryzatagline.cirad.fr/) integrates the phenotypic data obtained from the evaluation of Génoplante rice insertion line bank. In addition to the general data such as production records and seed stocks, the database also includes the GUS/GFP expression analysis of primary transformants (T0) and/or T1 progeny, as well as the morphological and physiological changes of T1 progeny detected in plant growth room or field agronomy. In the future, other phenotypic information will be integrated, including the expression of reporter genes in mature seeds, the changes of seed morphology and the response changes after inoculation with Magnaporthe grisea.
RiceRc Database
Rice Pan-genome database RiceRc (https://ricerc.sicau.edu.cn/) provides large-scale genome resources, including 171,072 structural variations (SV) and 25,549 genome copy number variations (gCNV). At the same time, it also integrates genome browser and BLAST function, which is convenient for users to query genome sequence and genetic variation, and promotes functional genomics and breeding research of rice. This database is helpful to identify phenotypic genetic variation that can not be detected by SNPs and single reference genome, thus providing important resources for rice breeding and plant functional genomics and evolutionary biology research.
RiceSuperPIRdb database
Rice Super-pan-genome information resource database RiceSuperPIRdb (http://www.ricesuperpir.com/) is a web-based interactive rice super-pan-genome information resource platform, which provides a variety of key data to support rice functional genomics and breeding research. Users can access the following data:
- Sequence and gene annotation of each assembly
- All assembled non-reference whole genome alignment
- Super Pan Genome Map and its related gene annotations
- A variation map for connecting the query data with the variation of the super pan-genome
- Notes of each assembled swivel component.
The database aims to promote the application of these research results, and will continuously update more data and analysis results.
RIGW Database
RIGW (http://rice.hzau.edu.cn/rice_rs3/), a comprehensive database of indica rice genome, is a comprehensive bioinformatics platform specially designed for rice research, which is mainly based on the genome data of Zhenshan 97 (ZS97) and Minghui 63 (MH63). This database provides a variety of data resources including genome, transcriptome, protein-protein interaction (PPI) and metabolic network. RIGW was upgraded to version 3.0 by assembling ZS97RS3 and MH63RS3 with the latest gap-free reference genome, and the sequenced genome and related information were displayed in a systematic and graphical way, which was convenient for researchers to conduct in-depth comparative research.
Conclusion
Generally, the landscape of rice research has been revolutionized by the multitude of databases available. These databases, such as RAP-DB, RGAP, and RGI, cover a wide spectrum of information, from genome sequences and annotations to gene expression profiles and mutant data. They play a crucial role in promoting rice genetics and breeding research. By integrating diverse data resources, researchers can identify genes related to traits like yield, nutritional quality, and environmental adaptability more efficiently. As technology advances and more data is generated, continuous updates to these databases will ensure they remain powerful tools. This will ultimately facilitate the development of sustainable rice varieties to meet the challenges of a growing global population and climate change.
References
- Shivute F.N, Zhong Yi., et al. "Genome-wide and pan-genomic analysis reveals rich variants of NBS-LRR genes in a newly developed wild rice line from Oryza alta Swallen." Frontiers in Plant Science 15 (2024): 1345708. https://doi.org/10.3389/fpls.2024.1345708
- Shang Lianguang, He Wenchuang., et al. "A complete assembly of the rice nipponbare reference genome" Molecular plant 16 (2023): 1232-1236. https://doi.org/10.1016/j.molp.2023.08.003
- Wang Jian, Yang Wu., et al. "A pangenome analysis pipeline provides insights into functional gene identification in rice." Genome Biology 24 (2023). https://doi.org/10.1101/2022.06.15.496234
- Zhou Yong, Yu Zhichao., et al. "Pan-genome inversion index reveals evolutionary insights into the subpopulation structure of asian rice." Nature Communications 14 (2023): 1567. https://doi.org/10.1038/s41467-023-37004-y
- Wu Dongya, Xie Lingjuan., et al. "A syntelog-based pan-genome provides insights into rice domestication and de-domestication." Genome Biology 24 (2023): 179. https://doi.org/10.1186/s13059-023-03017-5
- Woldegiorgis S T, Wu Ti., et al. "Identification of heat-tolerant genes in non-reference sequences in rice by integrating pan-genome, transcriptomics, and QTLs." Genes 13 (2022): 1353. https://doi.org/10.3390/genes13081353
- Singh P K, Rawal H C., et al. "Pan-genomic, transcriptomic, and mirna analyses to decipher genetic diversity and anthocyanin pathway genes among the traditional rice landraces." Genomics 5 (2022): 110436. http://dx.doi.org/10.1016/j.ygeno.2022.110436
- Zhang Fan, Xue Hongzhang., et al. "Long-read sequencing of 111 rice genomes reveals significantly larger pan-genomes." Genome Research 32 (2022): 853-863. https://doi.org/10.1101/gr.276015.121
- Shang Lianguang, Li Xiaoxia., et al. "A super pan-genomic landscape of rice." Cell Res 32 (2022): 878-896. https://doi.org/10.1038/s41422-022-00685-z
 Send a Message
Send a MessageFor any general inquiries, please fill out the form below.