Mechanisms of Plant Chloroplast Biogenesis
Photosynthesis is a fundamental biological process that sustains life on Earth, occurring within chloroplasts in eukaryotic cells. Chloroplasts are hypothesized to have originated from a symbiotic relationship between ancestral photosynthetic prokaryotes and heterotrophic eukaryotes more than a billion years ago. This endosymbiotic event marked a pivotal moment in evolutionary history, leading to the development of chloroplasts as essential organelles for photosynthesis.
Significance of Chloroplast Biogenesis in Crop Improvement
Enhancing photosynthetic efficiency remains a significant goal in crop improvement due to its potential impact on agricultural productivity. Chloroplast biogenesis, the process by which new chloroplasts are formed, is crucial for optimizing photosynthetic function. A comprehensive understanding of the regulatory mechanisms governing this process is essential for developing strategies to improve crop yields and sustainability.
Regulatory Mechanisms of Chloroplast Biogenesis
Chloroplast biogenesis is regulated by a complex network of transcription factors. Among these, the GOLDEN2-LIKE (GLK) family of transcription factors plays a central role. GLK transcription factors are pivotal in orchestrating the development of chloroplasts and ensuring their proper function. Despite their crucial role, the full spectrum of proteins involved in chloroplast biogenesis remains incompletely characterized.
Challenges in Identifying Regulatory Proteins
Research on glk mutants has revealed residual chlorophyll content, suggesting that additional proteins contribute to chloroplast biogenesis. However, the identification of these proteins has been impeded by the limitations of forward genetic approaches. Genetic redundancy has been proposed as a major obstacle, wherein multiple genes or proteins may perform overlapping functions, complicating their identification and characterization.
Case Study
Significant Research Advances in Chloroplast Biogenesis: MYB-Related Transcription Factors Uncovered
To address these challenges, researchers, including Julian M. Hibberd from the University of Cambridge, have advocated for the use of species with more compact genomes, such as Marchantia polymorpha. The analysis of such species may provide clearer insights into the regulatory mechanisms of chloroplast biogenesis by reducing genetic redundancy and simplifying the identification of key regulatory proteins.
On July 23, 2024, Cell published a groundbreaking study led by Julian M. Hibberd from the University of Cambridge. The article, titled "MYB-Related Transcription Factors Control Chloroplast Biogenesis," presents novel insights into the regulatory mechanisms governing chloroplast development and photosynthetic gene expression.
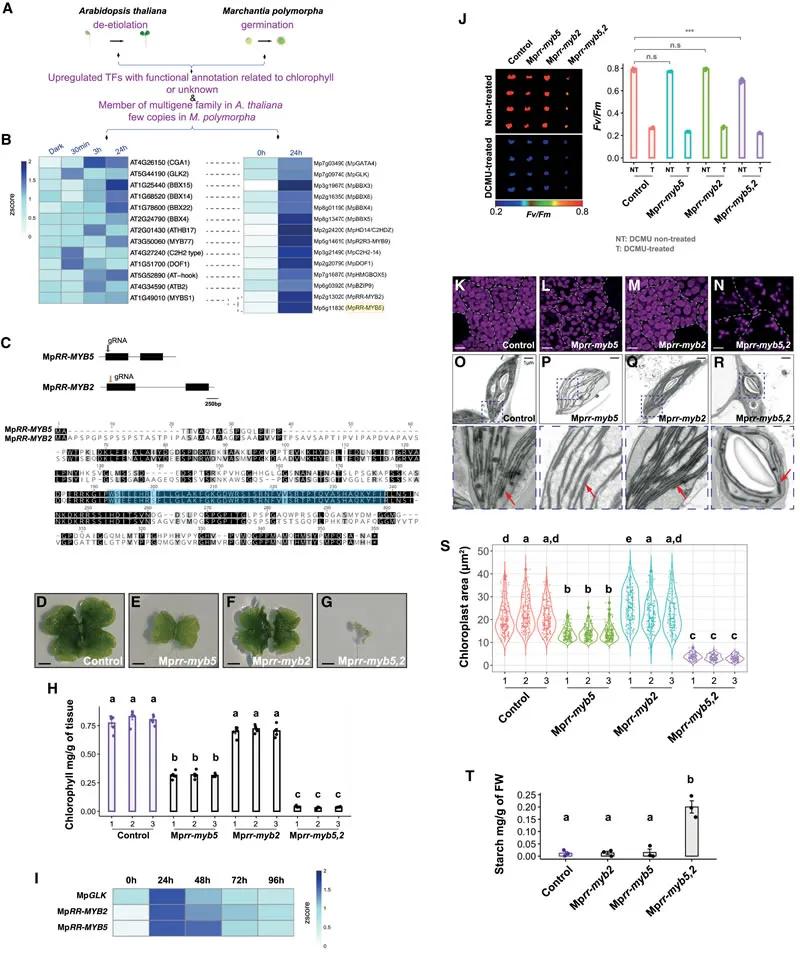 Fig. 1 MpRR-MYB5 and MpRR-MYB2 coordinately regulate chloroplast biogenesis
Fig. 1 MpRR-MYB5 and MpRR-MYB2 coordinately regulate chloroplast biogenesis
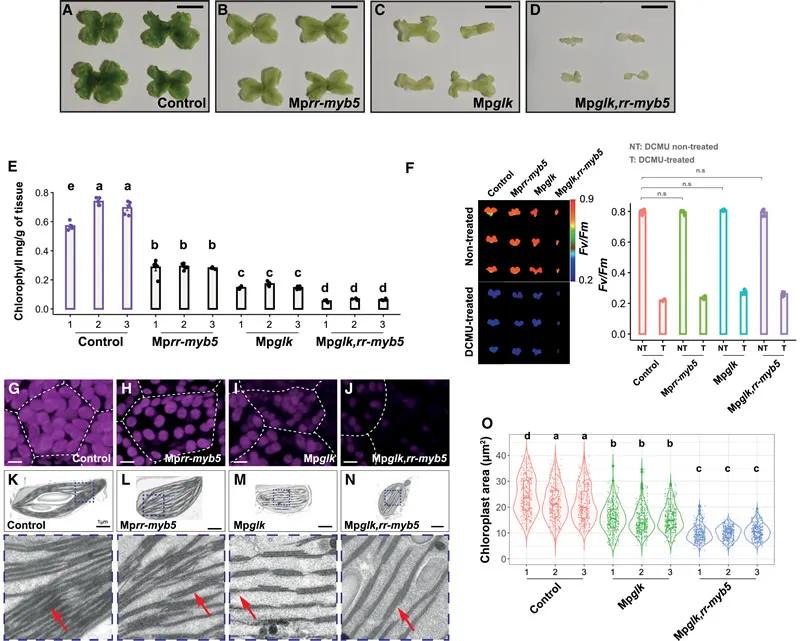 Fig. 2 MpRR-MYB5 cooperates with MpGLK to regulate chloroplast biogenesis
Fig. 2 MpRR-MYB5 cooperates with MpGLK to regulate chloroplast biogenesis
Introduction
The study employed reanalysis of publicly available RNA sequencing data, gene editing techniques, and phenotypic analysis to discover two RR-type MYB-related transcription factors, MpRR-MYB5 and MpRR-MYB2, which play pivotal roles in chloroplast biogenesis and photosynthetic gene expression.
Chloroplasts are vital organelles responsible for photosynthesis in eukaryotic cells. Understanding the regulatory mechanisms underlying chloroplast biogenesis is crucial for enhancing photosynthetic efficiency and improving crop yield. Recent research has focused on identifying key transcription factors involved in this process. MYB-related transcription factors, particularly those of the RR-type, have emerged as significant regulators of chloroplast development.
Methods
The research team utilized a multifaceted approach to identify transcription factors involved in chloroplast biogenesis:
Reanalysis of RNA Sequencing Data: Publicly available RNA sequencing datasets were reexamined to identify potential transcription factors associated with chloroplast development.
Gene Editing: Targeted gene editing was performed to create mutants for functional analysis.
Phenotypic Analysis: Phenotypic characteristics of the mutants were assessed to evaluate the impact on chloroplast biogenesis and photosynthetic gene expression.
Service you may interested in
Results
The study identified two RR-type MYB-related transcription factors, MpRR-MYB5 and MpRR-MYB2, as crucial regulators of chloroplast biogenesis and photosynthetic gene expression. The key findings are summarized below:
MpRR-MYB5 and MpRR-MYB2: Both transcription factors exhibit cooperative functions in chloroplast biogenesis. Double mutants, Mprr-myb5,2, displayed severely impaired chloroplast development, significantly reduced chlorophyll content, smaller chloroplasts, and underdeveloped thylakoid membranes.
Interaction with MpGLK: MpRR-MYB5 and MpRR-MYB2 work synergistically with MpGLK (GOLDEN2-LIKE) transcription factors. While MpGLK mutants also showed compromised chloroplast development, the severity was less pronounced compared to the Mprr-myb5,2 double mutants. In Mpglk,rr-myb5 double mutants, chloroplasts were smaller, thylakoid membranes were reduced, and granal stacking was diminished.
Gene Expression Regulation: MpRR-MYB5 and MpRR-MYB2 regulate the expression of genes involved in chlorophyll biosynthesis and photosynthesis. Mutations in these factors led to decreased transcript levels of relevant genes and reduced expression of photosynthesis-associated proteins.
DNA Affinity Purification and Sequencing (DAP-seq)
DAP-seq analysis revealed that MpRR-MYB5 and MpRR-MYB2 bind to the promoters of photosynthetic genes and activate their transcription. This finding underscores their role in regulating the expression of genes critical for chloroplast function.
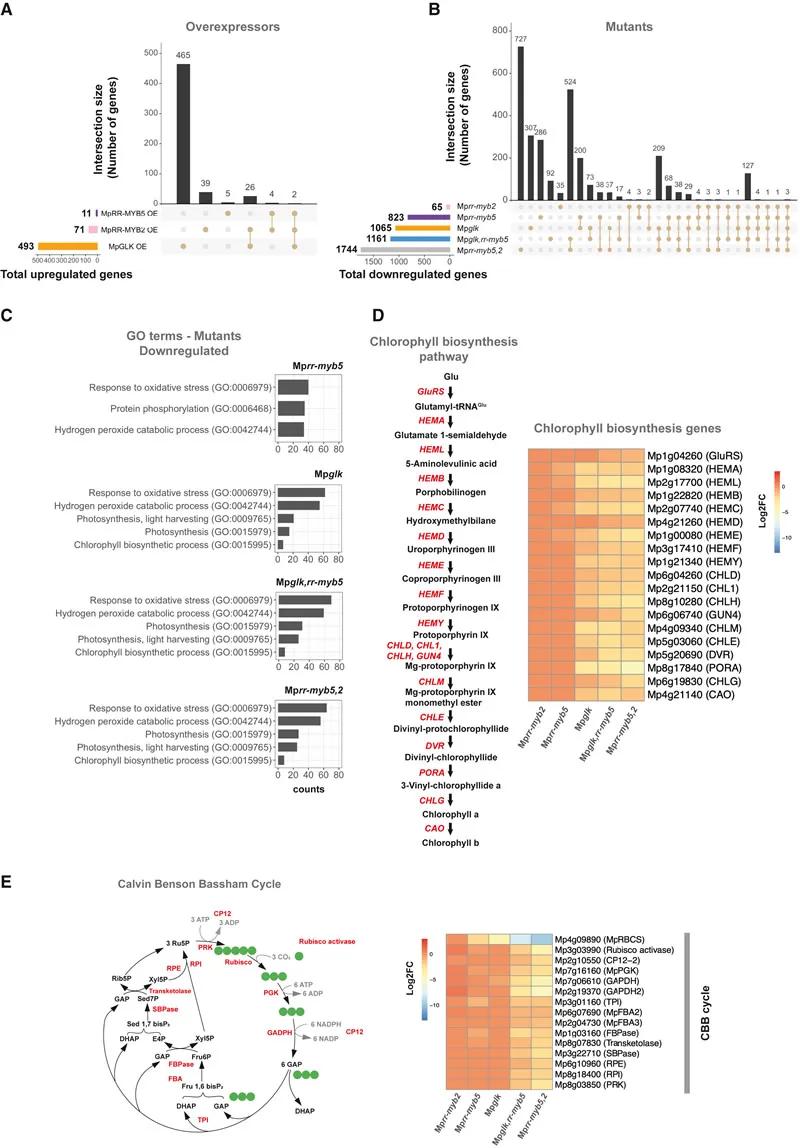 Fig. 3 MpRR-MYB5 and MpRR-MYB2 regulate the expression of genes encoding chlorophyll biosynthesis and the Calvin Benson cycle
Fig. 3 MpRR-MYB5 and MpRR-MYB2 regulate the expression of genes encoding chlorophyll biosynthesis and the Calvin Benson cycle
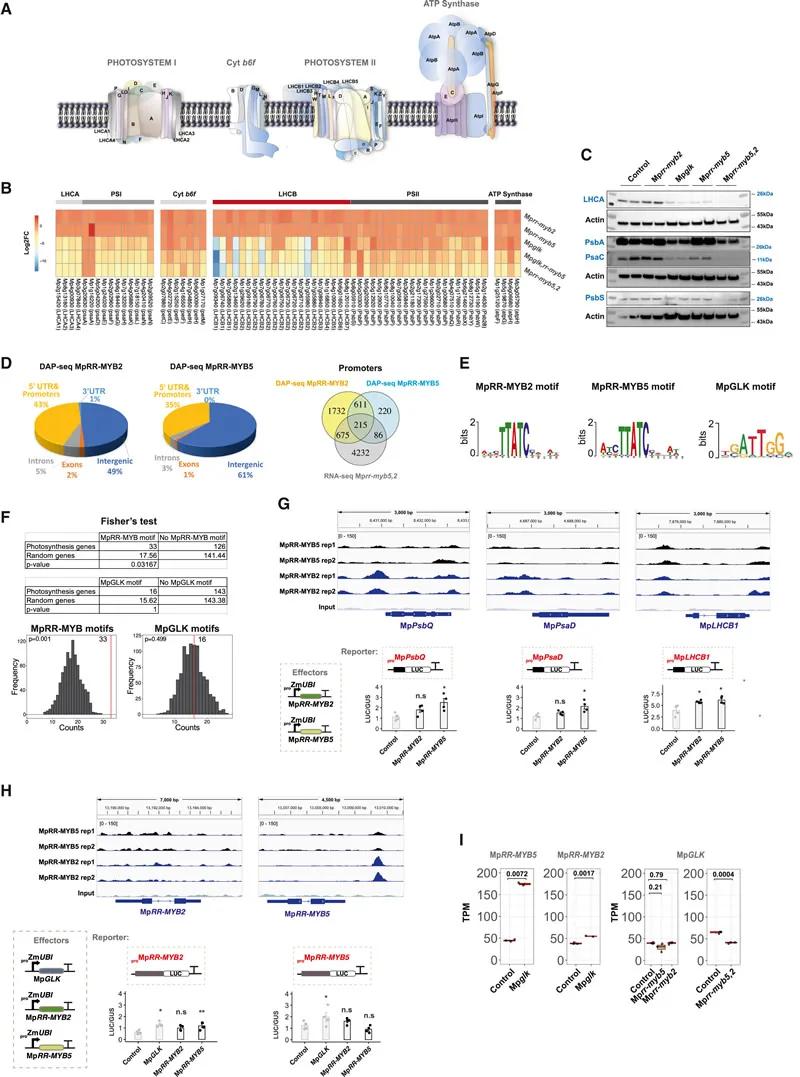 Fig. 4 MpRR-MYB5&2 targets photosynthesis genes
Fig. 4 MpRR-MYB5&2 targets photosynthesis genes
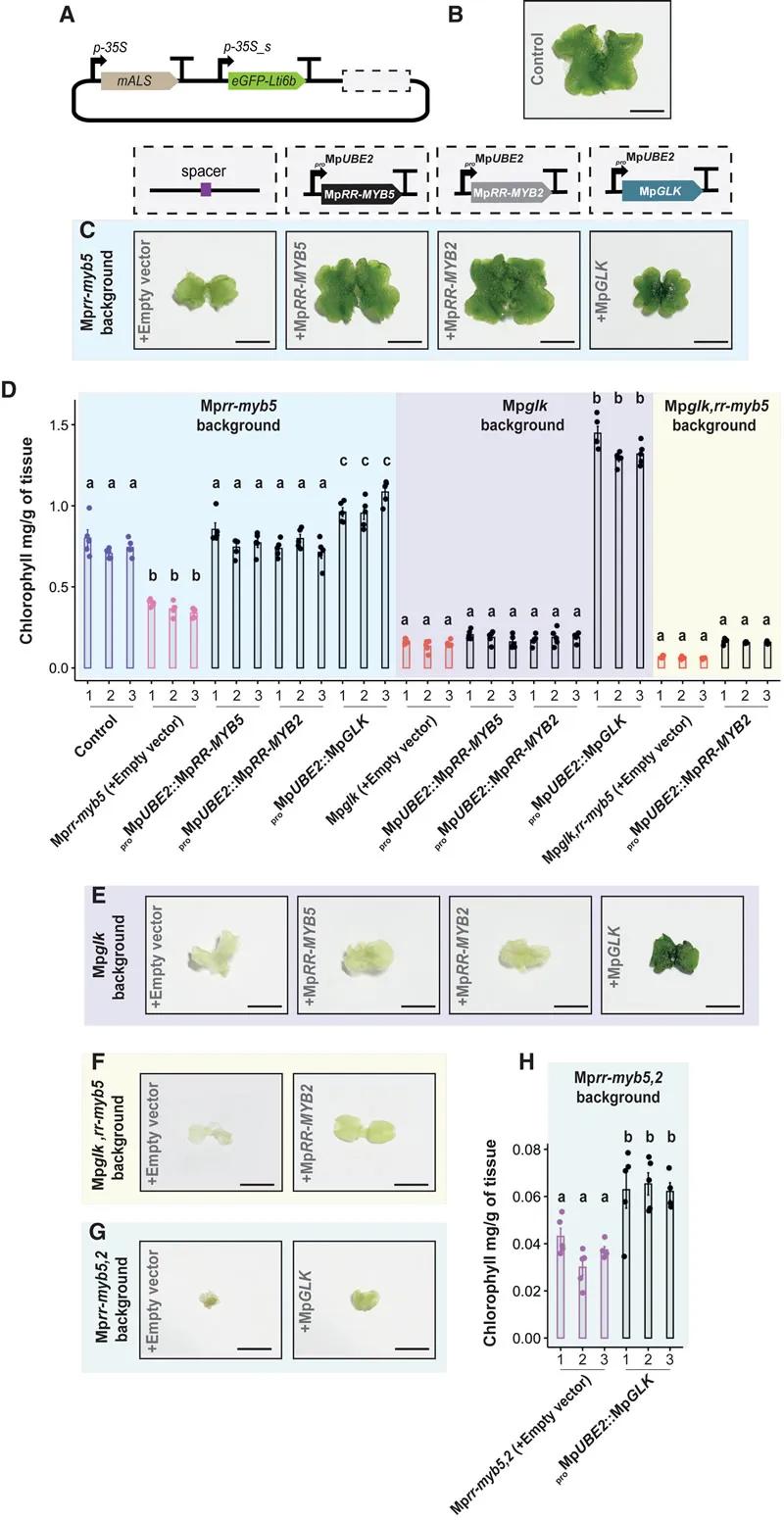 Fig. 5 RR-MYBs have mutual functional redundancy, but the redundancy between MpRR-MYBs and MpGLK is limited
Fig. 5 RR-MYBs have mutual functional redundancy, but the redundancy between MpRR-MYBs and MpGLK is limited
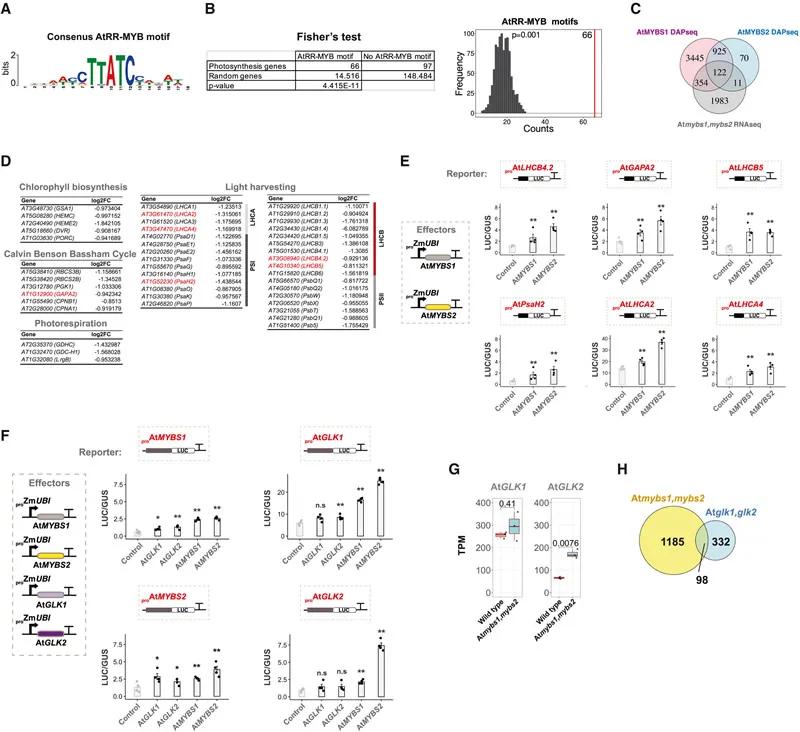 Fig. 6 AtMYBS1&2 can bind and activate photosynthesis genes and transcription factors involved in chloroplast biogenesis
Fig. 6 AtMYBS1&2 can bind and activate photosynthesis genes and transcription factors involved in chloroplast biogenesis
Impact and Future Directions: The Role of MYB-Related Transcription Factors in Chloroplast Biogenesis
The recent study on RR-MYBs has significant implications for the field of plant biology. By elucidating the role of these transcription factors in chloroplast biogenesis, the research offers new insights and directions for further investigation into chloroplast formation and function. The identification of RR-MYBs addresses a critical gap in the understanding of chloroplast developmental regulation and opens avenues for enhancing photosynthetic efficiency and crop yield through genetic and breeding approaches.
Implications for Plant Biology
Novel Insights into Chloroplast Development
The discovery of RR-MYBs contributes substantially to the knowledge of chloroplast biogenesis. These factors fill a crucial gap in the comprehension of the regulatory mechanisms governing chloroplast development. Their identification provides a fresh perspective on the formation and functionality of chloroplasts, offering a broader framework for studying these vital organelles. RR-MYBs have been shown to play a pivotal role in chloroplast biogenesis, with their target genes encompassing a wider range than those regulated by the previously known GOLDEN2-LIKE (GLK) transcription factors.
Potential for Genetic Improvement
The research highlights the potential of RR-MYBs as novel targets for genetic engineering and breeding strategies aimed at improving crop photosynthetic efficiency and yield. Given that RR-MYBs regulate a broader spectrum of genes involved in chloroplast development compared to GLK factors, they represent promising targets for developing crops with enhanced photosynthetic capabilities. This advancement could significantly contribute to crop improvement and agricultural progress, addressing global challenges related to food security and agricultural productivity.
Complex Interactions within Gene Regulatory Networks
The study reveals complex interactions and synergistic regulatory relationships between RR-MYBs and GLK transcription factors. This finding underscores the intricate nature of gene regulatory networks in plants and their response to environmental and developmental signals. The elucidation of these interactions will likely stimulate further research into the complexity of plant gene regulation and how these networks modulate physiological processes in response to various stimuli.
Evolutionary Conservation and Adaptation
From an evolutionary perspective, RR-MYBs are widely conserved among terrestrial plants and exhibit functional conservation across different species. This conservation provides valuable insights into how plants have adapted to environmental changes throughout evolution and developed complex physiological traits. Understanding the evolutionary roles of RR-MYBs can enhance our knowledge of plant adaptation mechanisms and evolutionary biology.
Broader Biological Connections
Linkage to Metabolic Processes
Chloroplast biogenesis is closely linked to various metabolic processes, including nitrogen and sulfur assimilation, amino acid and fatty acid biosynthesis, and carotenoid production. The study of RR-MYBs may yield new insights into these interconnected biological processes, contributing to a more comprehensive understanding of plant physiological metabolism. By exploring how RR-MYBs influence these processes, researchers can gain deeper insights into the overall physiological and metabolic functioning of plants.
Conclusion
The research on MYB-related transcription factors significantly advances the field of plant biology by providing new insights into chloroplast biogenesis and regulation. The identification of RR-MYBs fills a crucial knowledge gap and presents novel opportunities for enhancing crop productivity through genetic and breeding innovations. Furthermore, the complex interactions between RR-MYBs and GLK factors offer a deeper understanding of plant gene regulatory networks and their evolutionary significance. This study not only contributes to the fundamental understanding of plant biology but also holds promise for practical applications in agriculture and crop improvement.
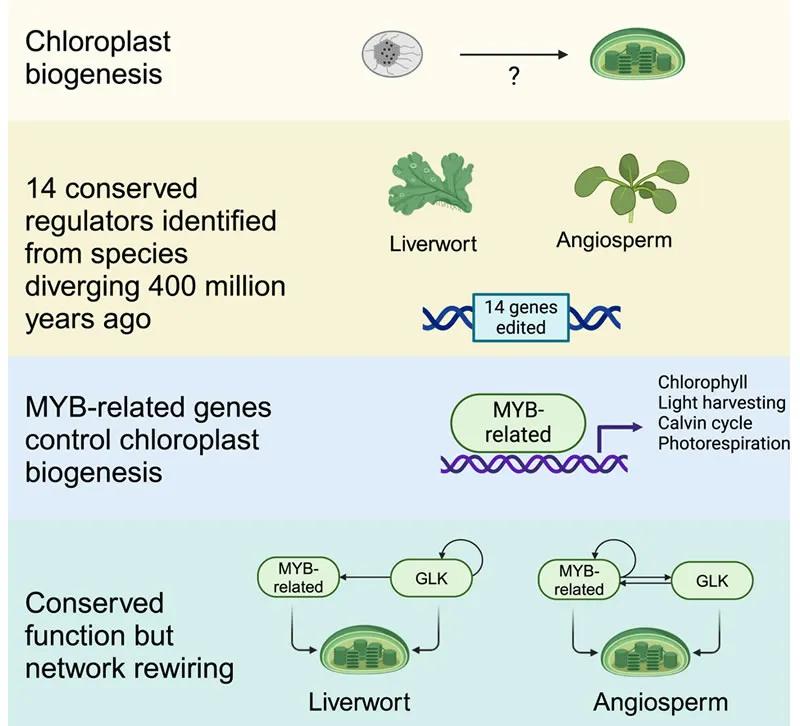
Future research must further elucidate the precise architecture of the gene regulatory network involving RR-MYBs and GLK. This includes defining the spatial and temporal distribution of their targets using methods such as ChIP-seq, understanding how these targets are regulated by developmental and environmental signals, and exploring the interactions of RR-MYBs with light signaling networks and hormone networks. Additionally, it is crucial to investigate their interactions with other proteins and post-translational modifications.
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.


 Fig. 1 MpRR-MYB5 and MpRR-MYB2 coordinately regulate chloroplast biogenesis
Fig. 1 MpRR-MYB5 and MpRR-MYB2 coordinately regulate chloroplast biogenesis Fig. 2 MpRR-MYB5 cooperates with MpGLK to regulate chloroplast biogenesis
Fig. 2 MpRR-MYB5 cooperates with MpGLK to regulate chloroplast biogenesis Fig. 3 MpRR-MYB5 and MpRR-MYB2 regulate the expression of genes encoding chlorophyll biosynthesis and the Calvin Benson cycle
Fig. 3 MpRR-MYB5 and MpRR-MYB2 regulate the expression of genes encoding chlorophyll biosynthesis and the Calvin Benson cycle Fig. 4 MpRR-MYB5&2 targets photosynthesis genes
Fig. 4 MpRR-MYB5&2 targets photosynthesis genes Fig. 5 RR-MYBs have mutual functional redundancy, but the redundancy between MpRR-MYBs and MpGLK is limited
Fig. 5 RR-MYBs have mutual functional redundancy, but the redundancy between MpRR-MYBs and MpGLK is limited Fig. 6 AtMYBS1&2 can bind and activate photosynthesis genes and transcription factors involved in chloroplast biogenesis
Fig. 6 AtMYBS1&2 can bind and activate photosynthesis genes and transcription factors involved in chloroplast biogenesis
