Human microbes are widely distributed in the gut, oral cavity, skin, bronchus, reproductive tract, organ tissues, and even in the blood, according to some researchers. In recent years, it has been found that gut microbes play an important role in all types of human diseases and immunity. With the advancement of sequencing technology, the study of microbes has also entered the era of omics; By analyzing the genetic material of microbial populations in the gut, researchers can identify the specific species and strains present, as well as their relative abundance and function. So how can microbiomics be used to study the effects of a particular type of disease or a certain condition of microbes? The following is a brief introduction to the application of microbiomics in the study of inflammation and disease.
16s amplicon sequencing resolves the relationship between gut microbes, blood cytokines and early miscarriage
One of the most important techniques used in gut microbiota research is 16S rRNA sequencing . This method involves sequencing a specific region of bacterial DNA that is highly conserved across different species. By comparing the sequences of these regions from different samples, researchers can identify the types of bacteria present and estimate their abundance.
It has been found that intestinal flora affects the host immune response and leads to imbalance in cytokine levels. In turn, dysregulation of the cytokine network is thought to be associated with the pathogenesis of unexplained miscarriage. Researchers investigated how gut microbial dysbiosis interferes with cellular immune function during miscarriage by 16s amplicon sequencing. Using sequencing and metabolomic data molecules, the authors found that several pathogens, such as Bacteroidales_S24_7_group and Eubacterium ruminantium_group, were significantly overrepresented in the feces of patients with miscarriage, while γ-amastigotes were the most overrepresented in the control group. This study highlights the network between gut microbiota, fecal metabolites, and Th1/Th17-mediated immune responses in patients with miscarriage.
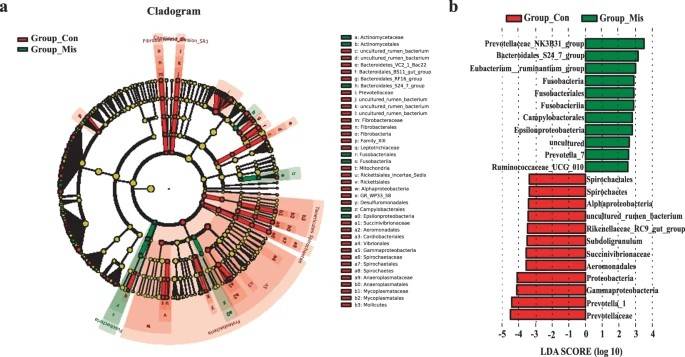 Compositions of different species in the control and miscarriage groups by LEfSe analyses
Compositions of different species in the control and miscarriage groups by LEfSe analyses
Metagenomic sequencing in combination with other omics reveals multiple gut microbe-metabolite-cytokine interrelationships in COVID-19-related complications
Another sequencing technique used in gut microbiota research is shotgun metagenomic sequencing. This method involves sequencing all of the DNA present in a sample, including both bacterial and human DNA. By analyzing this data, researchers can identify not only the types of bacteria present but also their functional capabilities, including their ability to produce metabolites that influence inflammation and other biological processes.
Cytokine storm is a major driver of COVID-19 pathogenesis. And several studies have shown that partial microbiota depletion is associated with the inflammatory response to disease. The authors attempted to find microbial-metabolite-cytokine interrelationships and new markers and therapeutic targets by performing shotgun metagenomic sequencing and metabolomic analysis of feces from 112 SARS-CoV-2 infected patients and control individuals matched for significant confounders, and cytokine measurements of plasma. The authors identified multiple gut microbe-metabolite-cytokine interrelationships in COVID-19-related complications, and in the future, it may be possible to predict COVID-19 and severe disease by testing the microbiome.
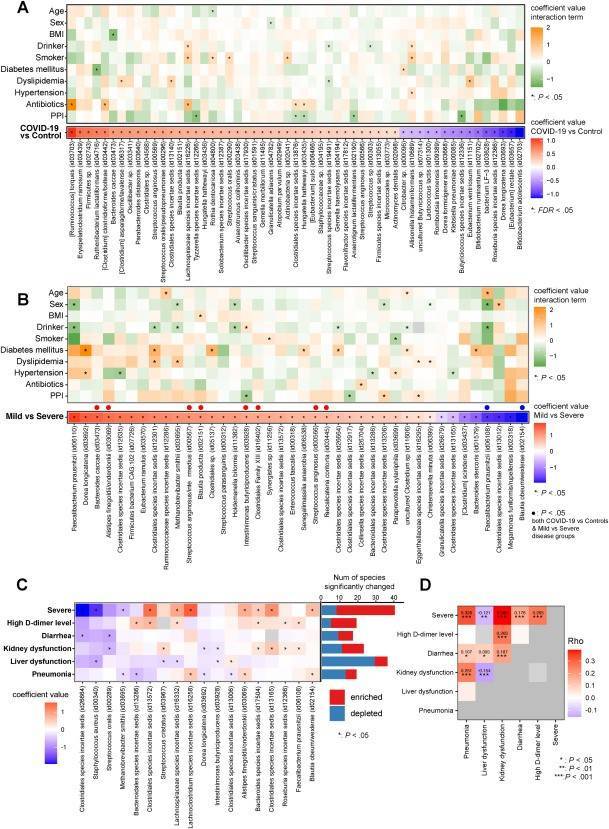 Distinct gut microbial variations in COVID-19 and background factors
Distinct gut microbial variations in COVID-19 and background factors
References:
- Liu Y, Chen H, Feng L, et al. Interactions between gut microbiota and metabolites modulate cytokine network imbalances in women with unexplained miscarriage. NPJ biofilms and microbiomes, 2021, 7(1): 1-12.
- Nagata N, Takeuchi T, Masuoka H, et al. Human gut microbiota and its metabolites impact immune responses in COVID-19 and its complications. Gastroenterology, 2023, 164(2): 272-288.
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.


 Sample Submission Guidelines
Sample Submission Guidelines
 Compositions of different species in the control and miscarriage groups by LEfSe analyses
Compositions of different species in the control and miscarriage groups by LEfSe analyses Distinct gut microbial variations in COVID-19 and background factors
Distinct gut microbial variations in COVID-19 and background factors