We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy


We are dedicated to providing outstanding customer service and being reachable at all times.
Long-read Genome Sequencing of Bats
At a glance:
- Overview
- Advantages of Long-read Genome Sequencing in Bats
- Applications of Long-read Genome Sequencing in Bats
Overview
With more than 1,400 species identified to date, bats (Chiroptera) account for about 20% of all extant mammal species. bats are found throughout the world and successfully occupy different ecological niches. From the smallest bumblebee bat (Craseonycteris thonglongyai) to the large (1 kilogram) golden-topped fruit bat (Acerodon jubatus), they possess extraordinary adaptations, including flight, echolocation, extremely long lifespans, and a unique immunity that both fascinates and frightens people. Over the past two decades, bats have become an important model system for studying host-pathogen interactions. In addition, bats can serve as excellent new models for studying aging, inflammation, and cancer, as well as other important biological processes. High-quality genomes are critical for understanding the molecular basis and evolution of unique traits in bats.
Now, an international consortium of bat biologists, computational scientists, conservation organizations, and genome technologists has set out to decode the genomes of all 1,300 species of bats using the PacBio SMRT Sequencing Technology and other technologies (Oxford Nanopore Sequencing Technology). The large-scale sequencing project will be completed in three phases, first sequencing 21 representatives of each bat family, then 220 representatives of each bat genus, and finally sequencing the remaining 1,288 species. It will greatly expand the 14 bat genome assemblies currently available in the National Center for Biotechnology Information (NCBI) database, which vary in quality and completeness.
Traditional sequencing methods, while effective, often face challenges in comprehensively capturing intricate genomes. The advent of long-read sequencing provides a fine-grained perspective that allows for a more detailed exploration of the bat genome. A reference-quality bat genome based on long-read sequencing could provide the resources needed to reveal and validate the genomic basis of bat adaptation and inspire new avenues of research directly related to human health and disease.
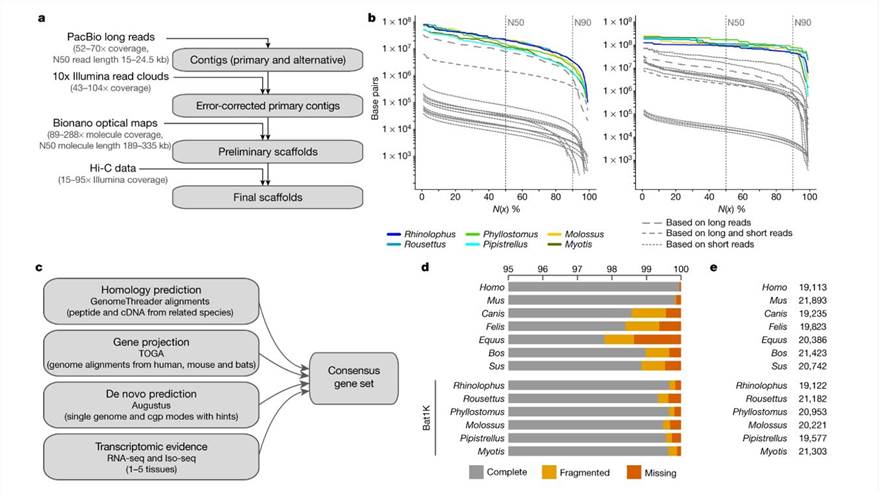 Assembly and annotation of the genomes of six bat species. (Jebb et al., 2020)
Assembly and annotation of the genomes of six bat species. (Jebb et al., 2020)
Advantages of Long-read Genome Sequencing in Bats
Precision and Accuracy
One of the paramount advantages of long-read sequencing is its high-fidelity genomic capture. When we observe chiropteran genomic data, it becomes crucial to scrutinize intricate genomic segments, especially those harboring duplicated gene clusters. Traditional short-read sequencing modalities might inadvertently elide these regions, thereby leading to a truncated capture of pivotal genetic blueprints.
Unrivaled Insight into Evolutionary Traits
The order Chiroptera manifests an array of evolutionary marvels, ranging from advanced laryngeal echolocation mechanisms to sophisticated and sometimes enigmatic immune response pathways. Harnessing the prowess of long-read sequencing enables us to microscopically examine the genomic underpinnings of these evolutionary adaptations. For instance, contemporary research deploying long-read sequencing paradigms has unveiled the positive selection pressure on auditory-centric genes in bats, signifying a protracted evolutionary journey toward echolocation proficiency.
Enhanced Gene Annotation
In the realm of genome research, gene annotation functions as the linchpin, illuminating both the functional dynamics and spatial location of genes. By synergistically amalgamating computational tools, such as the Tool for Inferring Homologs from Genome Alignment (TOGA), with long-read sequencing, we can augment the cartography of the chiropteran genome. This results in more exhaustive and punctilious gene annotations. For instance, comprehensive genome-wide screens have underscored genomic shifts potentially underpinning bats' unparalleled immunological prowess.
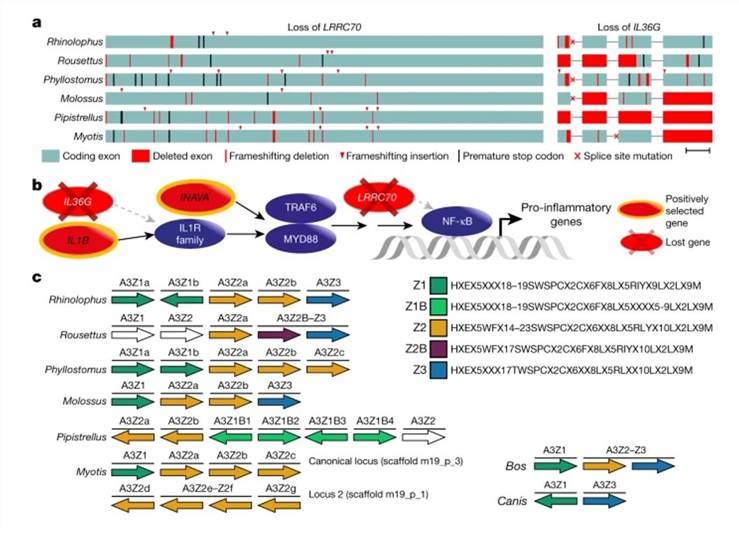 Genome-wide screens highlight changes in genes that are potentially involved in exceptional immunity in bats. (Jebb et al., 2020)
Genome-wide screens highlight changes in genes that are potentially involved in exceptional immunity in bats. (Jebb et al., 2020)
Applications of Long-read Genome Sequencing in Bats
Understanding Bat Immunity
The Chiroptera order plays host to myriad zoonotic viruses such as Ebola, SARS, and MERS, yet intriguingly, they display an asymptomatic phenotype and are resilient to the pathogenic onslaught. By harnessing long-read genome sequencing, we can embark on a deeper exploration into the chiropteran immune arsenal. This potentially reveals genetic pathways signifying the selective augmentation or diminution of immune-centric genes and the amplification of antiviral genetic circuits. Such revelations harbor profound implications for understanding human pathogenesis and infectious disease response mechanisms.In the face of escalating anthropogenic threats, bats necessitate targeted conservation strategies. By obtaining high-resolution genomes, we can formulate informed conservation paradigms and pinpoint species or populations teetering on the brink of endangerment. The Bat1K Initiative, for instance, aspires to harness sequencing technologies to encapsulate the genetic heterogeneity across bat species, thus fortifying our conservation arsenal.
Insights into Mammalian Evolution
Occupying a distinct phylogenetic niche, bats offer a lens into evolutionary dynamics not just limited to their order but spanning Mammalia. Through meticulous long-read genome sequencing, we stand on the precipice of delineating their intricate evolutionary pathways, enriching our comprehension of mammalian evolutionary dynamics.
Relevance to Human Health
Beyond the immediate understanding of chiropteran biology, genomic revelations have profound translational relevance. Elucidating genes implicated in bat longevity could potentially offer insights into human senescence. Similarly, demystifying chiropteran viral resilience could pave the way for innovative human strategies in infectious disease mitigation and management.
Reference
- Jebb, David, et al. "Six reference-quality genomes reveal evolution of bat adaptations." Nature 583.7817 (2020): 578-584.
Related Services
PacBio SMRT Sequencing Technology
Oxford Nanopore Sequencing Technology
For research purposes only, not intended for personal diagnosis, clinical testing, or health assessment