We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy


We are dedicated to providing outstanding customer service and being reachable at all times.
Conservation Genomics
At a glance:
- The Importance of Conservation Genomics
- Genomic Approaches in Biodiversity Research
- The Critical Role of Reference Genomes in Biodiversity Conservation
- Conservation Genomics Workflow to Guide Practical Management Actions
Conservation genomics is an emerging interdisciplinary field that harnesses the power of genomics to gain insights into biodiversity, species differentiation, and the evolutionary processes that influence species survival. It centers on the use of genomic data and technology to conserve the planet's biodiversity. By utilizing cutting-edge genomic technologies, natural resource conservationists can decode the genetic structure of species and elucidate patterns and processes that may have been overlooked by traditional ecological approaches.
The Importance of Conservation Genomics
The world is currently undergoing a critical phase, often referred to as the "sixth mass extinction". As ecosystems are threatened and species face extinction, traditional conservation approaches have proven inadequate. The value of incorporating genomics into conservation is manifold:
- High-resolution identification: traditional methods, such as phenotyping, provide only surface-level insights. Genomics delves deeper and can distinguish between subpopulations or even individual members of a species.
- Scientific decision-making: Public genomic resources greatly assist biodiversity conservation by providing concrete evidence for informed management decisions. By investigating a variety of genomic applications, conservationists can make timely and effective data-driven decisions.
- Monitoring and restoration: Genomics can monitor changes in genetic diversity over time, providing early warning signs of population decline or genetic bottlenecks. It also plays a vital role in restoration efforts by ensuring the genetic health of reintroduced or supported populations.
Genomic Approaches in Biodiversity Research
Conservation genomics has become a focal point for biodiversity research, harnessing the power of genomic data to inform conservation efforts. High-throughput genome sequencing technologies have recently evolved from primarily generating short (50-300 bp) DNA sequencing reads to longer (> 10 000 bp) DNA sequencing reads. Genomics has revolutionized biodiversity research through a range of technologies, each unlocking unique insights.
DNA Barcodes and Metabarcodes
DNA barcoding has reshaped the way we identify and monitor biodiversity. By targeting specific DNA loci surrounded by conserved flanking regions, this approach provides a unique genetic fingerprint for a wide range of organisms. Originally initiated for animals that use mitochondrial COI genes, it has since expanded to include vertebrates, plants, fungi, and even bacteria.
Metabarcoding is an advanced form of DNA barcoding that incorporates next-generation sequencing to evaluate mixed samples. This approach has been applied in different fields, such as tracking species turnover during ecosystem restoration or pinpointing invasive species. Notably, metabarcoding goes beyond traditional barcoding by allowing bulk sampling, thus increasing the throughput of biodiversity assessment.
However, there is one major limitation. Short DNA regions in barcodes sometimes fail to distinguish between closely related taxa or taxa with infiltrating genes. This is where genome skimming can provide a more refined approach.
Reducing Genomic Representation
The next advance in genomic tools is simplified representative DNA sequencing (RRS). Tailored for non-model species, RRS focuses on a limited portion of the genome in each sample. Although only this portion is captured, the data retrieved is reliable, providing researchers with information on genetic diversity, inbreeding coefficients, population structure, and more. Technologies such as RADseq or HyRAD are popular examples of RRS methods.
However, RRS is not without its flaws. For example, if different studies wish to combine their RRS-derived data, they must ensure that the same experimental protocols are used, a requirement that sometimes limits broader applications.
Gene Expression
Another component of conservation genomics is understanding gene expression, primarily through RNA-Seq. These data reveal functional variation in natural populations and provide insights into how species respond to environmental change. Recent studies have used gene expression data to link life history traits to population dynamics and even measure responses to pesticide exposure.
However, the complexity of RNA, which is susceptible to degradation and influenced by a variety of biological factors, means that gene expression studies require careful planning and interpretation.
As the field of conservation genomics matures, epigenomics is receiving increasing attention. Epigenetic modifications can lead to changes in gene expression without altering the DNA sequence, with far-reaching consequences for species adaptation and survival. For example, epigenetic changes may play a decisive role in how species respond to rapid environmental change.
Whole Genome Sequencing
Whole genome sequencing (WGS) is the most comprehensive tool in conservation genomics. It provides a holistic view of an organism's genetic makeup, allowing in-depth analysis of population history, genetic structure, and areas of natural selection. Whole genome sequencing allows researchers to identify rare genetic variations or mutations in regulatory elements that may affect species survival or adaptation.
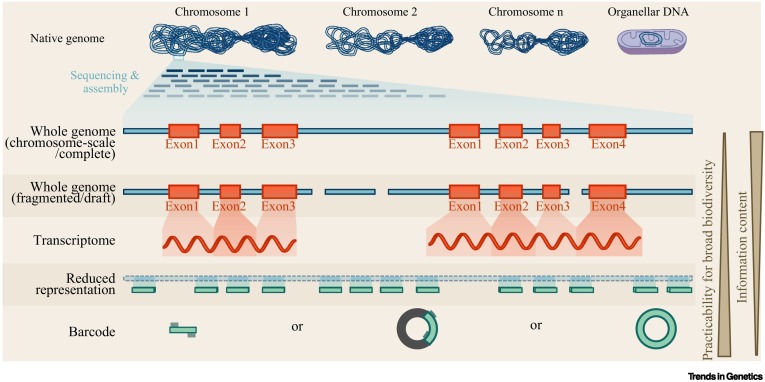 Genomic approaches for biodiversity research. (Theissinger et al., 2023)
Genomic approaches for biodiversity research. (Theissinger et al., 2023)
The Critical Role of Reference Genomes in Biodiversity Conservation
Reference genomes, epitomized by their superlative contiguity, unerring accuracy, and meticulously annotated genomic assemblies, serve as lynchpins in the sphere of intricate genomic research. Envisioning a reference genome, one could delineate it as a comprehensive cartographic representation of the genomic architecture inherent to a species, encompassing its multifaceted structural intricacies and organizational modalities. Mirroring the quintessence of model specimens within the realm of taxonomic sciences, the reference genome emerges as an archetypal paradigm, laying the groundwork for successive genomic inquiries.
Yet, a glaring limitation remains: the applicability of reference genomes is predominantly tethered to a select coterie of model organisms. However, an amalgamation of advanced sequencing modalities—be it the single-molecule long-read sequencing platforms like SMRT or nanopore sequencing or the use of linked reads typified by TELL-seq or stLFR for contig construction, augmented by optical maps, and/or proximity ligation reads (exemplified by 3C-seq or Hi-C) for adept scaffolding—heralds a universally applicable strategy. This strategy is primed to craft phased, chromosome-scale reference genomes that span the vast phylogenetic expanse encapsulating the Tree of Life.
Driving this monumental shift is a confluence of factors: the plummeting costs juxtaposed with the escalating scalability and fidelity of sequencing technologies; the emergence of sophisticated algorithmic tools; and quantum leaps in computational prowess. Collectively, these advancements have catalyzed the generation of reference genomes that straddle the rich tapestry of global biodiversity. Given their inherent profundity and precision, it's unequivocally evident that reference genomes stand at the vanguard of conservation genomics, illuminating pathways to judiciously shepherd our planet's precious genetic reservoirs.
Mapping Genome Diversity
The primary application of reference genomes in conservation genomics is to map genetic diversity within and between species. These genomes enable researchers to meticulously map and annotate this diversity. For example, they elucidate complex details such as patterns of linkage disequilibrium, structural variation, and even the integration or deletion of mobile genetic elements. In the field of conservation, where every nuance of genetic diversity can be a cornerstone of survival, this mapping is invaluable.
Uncovering Functional and Adaptive Genetic Variation
The central goal of biodiversity conservation is not only to protect species but also their inherent genetic strengths and potential. Reference genomes can help identify functional genetic variation underpinning important phenotypic traits. For example, through genome-wide association studies (GWAS), reference genomes highlight traits that are critical for resilience to anthropogenic threats such as climate change. Such identification is critical for initiatives such as captive breeding or translocation programs.
Deciphering the Mysteries of Inbreeding and Genetic Loading
In conservation genetics, inbreeding - the result of mating between closely related individuals - has always been a concern. With the precision of annotated reference genomes, understanding the complexities of inbreeding has become more feasible. These genomes reveal the genetic architecture of inbreeding depression, revealing important details such as the number of loci involved, the role of deleterious recessive alleles, and the dynamics of these alleles within populations. These insights have revolutionized translocation conservation management strategies.
Understanding Telecrossing and Gene Penetrance
Genomic dances between different genetic lineages sometimes lead to teleost decline. As a genomic compass, reference genomes guide us through the myriad complexities of genetic divergence, epistatic interactions, and the myriad complexities generated by human-mediated hybridization. They are a gateway to understanding the delicate balance of genetic introgression and its impact on local populations.
Unlocking the Potential for Adaptation
In an era of rapid environmental change, understanding adaptive potential is critical. With the reference genomes in our arsenal, we can identify variants adapted to local conditions that play a key role in ensuring the survival and prosperity of species. Recognizing these variants can guide conservation strategies and ensure that our efforts are aligned with the evolutionary potential of organisms.
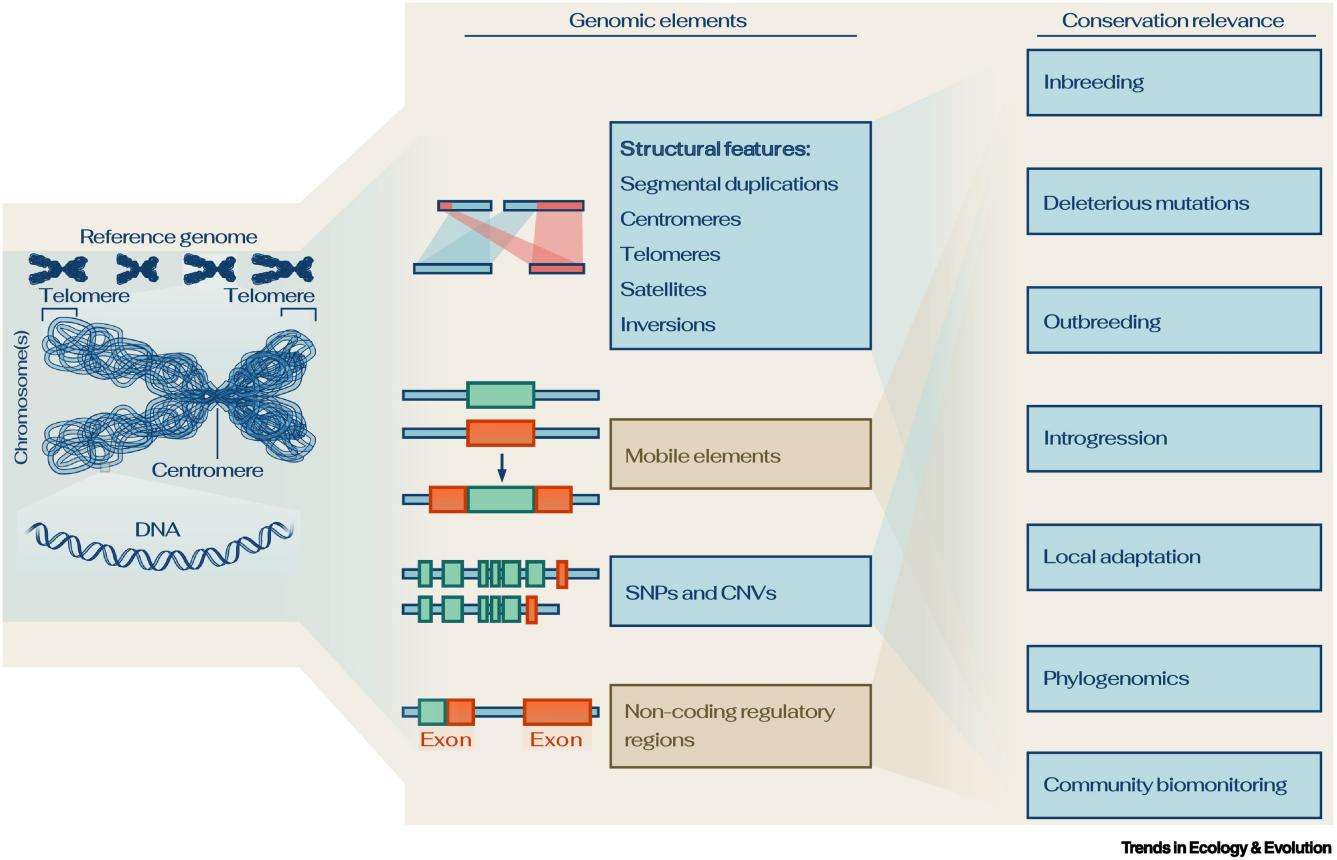 Reference genomes offer an (almost) complete record of the genome of a species. (Maumus et al., 2022)
Reference genomes offer an (almost) complete record of the genome of a species. (Maumus et al., 2022)
Conservation Genomics Workflow to Guide Practical Management Actions
(1) Sample collection and standardization
At the nexus of conservation genomics is the non-negotiable requirement for a meticulously designed and universally standardized sampling regime. Depending on the investigational ambit—whether delving into geographically circumscribed species or encompassing wider taxonomic entities—there exists an imperative to achieve a comprehensive and representative sampling that encapsulates the entire geographical and ecological expanse of the entity under study. Marrying the sophisticated paradigms of conservation biology with the nuanced tenets of population genetics furnishes the scaffolding for devising a sampling blueprint. The emphasis here pivots not merely on the volumetric magnitude of the samples but vitally hinges on their qualitative integrity and the granularity of their representativeness.
(2) High-throughput sequencing and long-read sequencing
The epoch of next-generation sequencing (NGS) and long-read sequencing has ushered a transformative era for conservation genomics. These avant-garde technological conduits empower conservationists to delve into the intricate recesses of the genome, even from the most diminutive of tissue specimens. Sequencing genotyping, as an illustrative case in point, has emerged as a financially judicious avenue, proffering a deluge of genomic elucidation. Such advancements facilitate an unprecedented granular comprehension of species' genetic architectures.
(3) Data analysis and interpretation
In today's data-drenched genomics milieu, sophisticated bioinformatics suites and algorithmic solutions have risen to the fore as quintessential tools. Tasked with the herculean mission of trawling voluminous genomic datasets, these tools unveil discernible genomic patterns and cryptic genetic signals. Such profound explorations elucidate the extent of genetic heterogeneity within and across populations, thereby illuminating pivotal insights encompassing reproductive viability, adaptive resilience, and the overarching evolutionary trajectory.
(4) Practical applications of genomic insights
Genomic elucidations, rich in depth and specificity, lay the foundation for crafting bespoke conservation interventions. For instance, the demarcation of regions imbued with heightened genetic diversity can direct conservation efforts towards these genetic reservoirs, accentuating their preservation. In a parallel vein, a nuanced understanding of interspecies hybridization corridors or the identification of predominantly clonal population patches can sharpen the precision of species reintroduction or translocation methodologies.
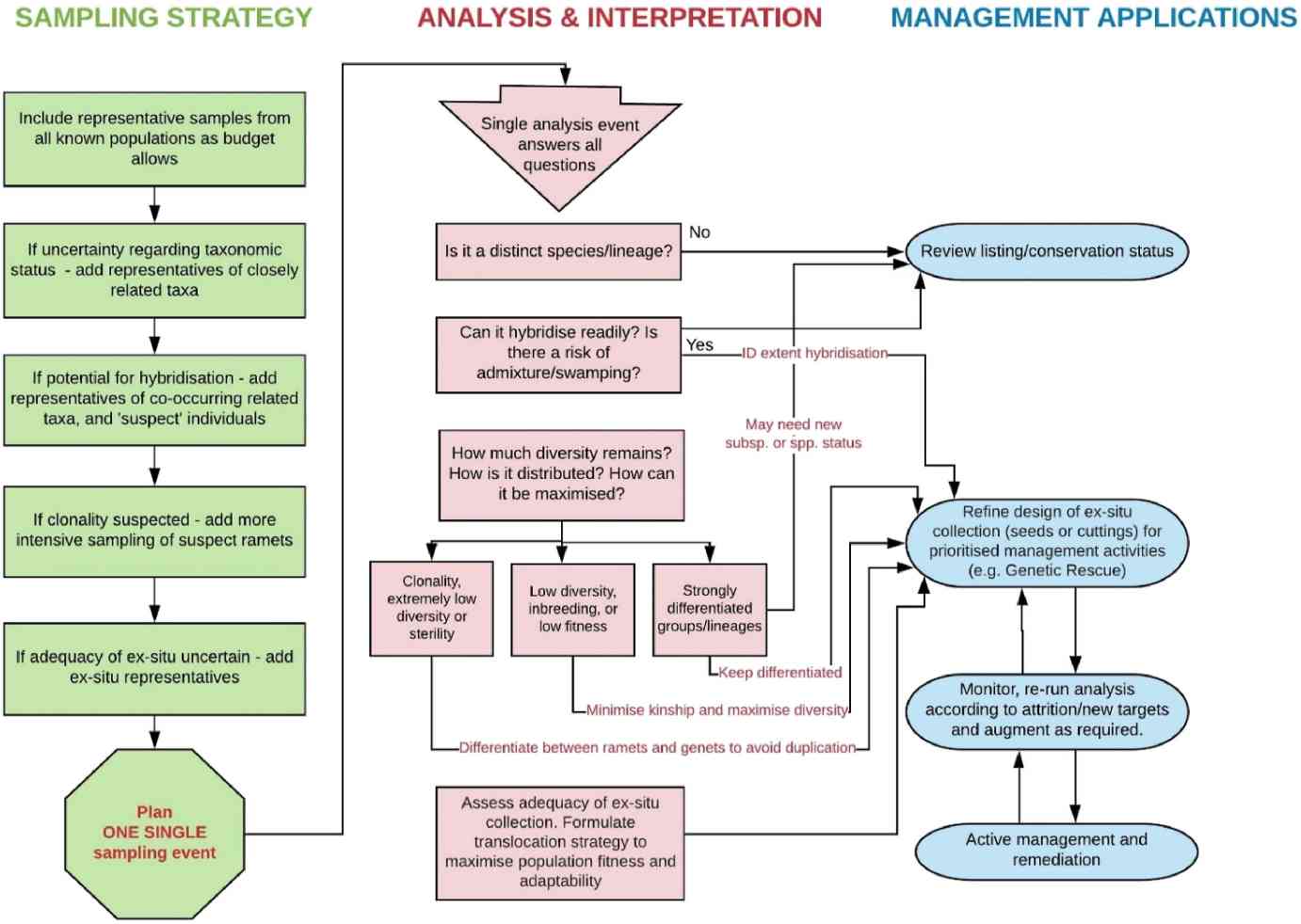 A simple design for an effective conservation genomic workflow. (Rossetto et al., 2021)
A simple design for an effective conservation genomic workflow. (Rossetto et al., 2021)
References
- Theissinger, Kathrin, et al. "How genomics can help biodiversity conservation." Trends in Genetics (2023).
- Maumus, Florian, et al. "The era of reference genomes in conservation genomics." (2022).
- Rossetto, Maurizio, et al. "A conservation genomics workflow to guide practical management actions." Global ecology and conservation 26 (2021): e01492.
Related Services
PacBio SMRT Sequencing Technology
Oxford Nanopore Sequencing
Whole-Genome Resequencing with Long-Read Sequencing
For research purposes only, not intended for personal diagnosis, clinical testing, or health assessment