We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy


We are dedicated to providing outstanding customer service and being reachable at all times.
Long-read Sequencing for DNA 5-methylcytosine (5mC) Analysis
At a glance:
- Overview
- PacBio Sequencing for 5mC Analysis
- Nanopore Sequencing for 5mC Analysis
- Comparison of PacBio and Nanopore Sequencing in 5mC Analysis
5-methylcytosine (5mC) is the most common form of DNA methylation and is involved in regulating many biological processes. Recent long-read length sequencing technologies, including Oxford Nanopore sequencing and PacBio HiFi sequencing, have dramatically expanded the ability to detect long-range, single-molecule, and direct DNA modifications without the need for additional laboratory techniques. CD Genomics is dedicated to providing a comprehensive elucidation of DNA methylation, providing valuable information on gene expression regulation at the chromosomal level.
Overview
DNA methylation is a fundamental epigenetic modification process that regulates gene expression and cellular responses to stimuli. Several different types of methylation, such as 5-methylcytosine (5mC), 5-hydroxymethylcytosine (5hmC), and n6 -methyladenosine (6mA), depending on the position of the atom in adenine or cytosine that is being modified, are among the most widely studied epigenetic modifications and play an important role in genome imprinting, regulation of chromatin structure, transposon inactivation, stem cell pluripotency and differentiation, inflammation, and regulation of transcriptional repression. play important roles. In addition, methylation in gene promoters affects transcriptional activity, and methylation in gene bodies affects silencing and selective splicing of repetitive DNA elements.
One of the most prevalent forms of DNA methylation in humans is 5mC, which usually occurs in the context of the CpG dinucleotide clustered into regions, known as CpG islands. DNA CpG methylation is an important epigenetic modification involved in the regulation of transcription, development, and genome stability. Loss of CpG modification is linked to cancer and inherited disorders such as Albright hereditary osteodystrophy (AHO), Beckwith-Wiedemann, Prader-Willi, and Angelman syndromes, and the development of the disease. and Angelman syndromes, and pseudohypoparathyroidism (PhP). Bisulfite sequencing (BS-seq) and its derivatives have become the gold standard for base-level resolution and quantitative 5mC analysis. However, this harsh chemical treatment degrades much of the DNA and produces sequencing libraries of low complexity. In addition, the short read lengths and systematic GC bias of short-read sequencing make it difficult to map read segments of complex repetitive genomic regions and to predict highly similar levels of methylated regions. It is estimated that approximately 10% of CpG sites in the human genome cannot be accurately localized after bisulfite modification using short-read-length technology.
In recent years, long-read sequencing technologies such as single-molecule real-time (SMRT) sequencing by Pacific Biosciences (PacBio) and nanopore sequencing by Oxford Nanopore Technologies (ONT) have emerged as emerging breakthroughs in the bottleneck of short-read segments for 5mC analysis due to their unprecedentedly long read-segment lengths (averaging up to 10kbp). They allow direct sequencing of natural DNA without PCR amplification and have been used for extensive analysis of genome methylation.
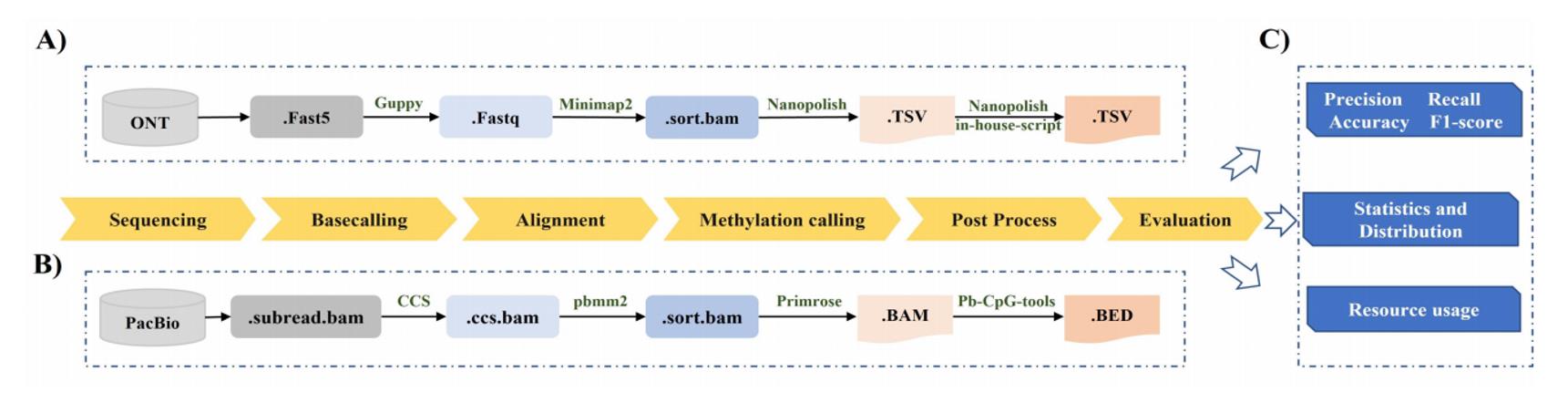 The pipelines of methylation calling for Nanopore and PacBio HiFi sequencing technologies, and the benchmark strategy. (Liu et al., 2022)
The pipelines of methylation calling for Nanopore and PacBio HiFi sequencing technologies, and the benchmark strategy. (Liu et al., 2022)
PacBio Sequencing for 5mC Analysis
PacBio Sequencing utilizes the principle that the presence of DNA modifications during SMRT sequencing affects DNA polymerase kinetics to directly detect DNA modifications. Recently introduced highly accurate long-read sequencing (HiFi reads) is generated using multiple cycles of consensus sequencing of long (up to ~25 kb) individual molecules.PacBio HiFi sequencing provides real-time observation of the polymerase as it binds fluorescently labeled nucleotides to synthesize DNA strands. Kinetic features including pulse width and pulse interval time correlate with chemical modifications of typical DNA bases, including 5mC modifications without bisulfite treatment. In summary, the PacBio HiFi read detects genome-wide 5mC and uses Primrose to identify allele-specific methylation at high resolution.
Recent advances have paved the way for dedicated tools optimized for PacBio sequencing data. A prominent example is "ccsmeth," a deep learning method developed specifically for the detection of DNA 5mCpG using PacBio's CCS reads. ccsmeth employs state-of-the-art neural network models, such as bi-directionally gated recurrent units (GRUs) and attentional mechanisms, to detect DNA 5mCpG at both the read level and genome-wide loci. ccsmeth uses state-of-the-art neural network models such as bi-directional gated recurrent units (GRUs) and attentional mechanisms to demonstrate superior accuracy in read-level and genome-wide site-level 5mCpG detection. Tools such as ccsmeth amplify the potential of PacBio sequencing to provide fine granularity for epigenomic studies.
Limitations of PacBio Sequencing:
SMRT sequencing can detect 5mC modifications at 250-fold coverage based on polymerase kinetics. However, this detection is not the result of direct 5mC detection at single-molecule resolution, but rather the aggregation of the subtle effects of 5mC on the kinetic signal of the polymerase during DNA synthesis. Thus, the requirement for high coverage of SMRT and the inability to directly detect single-molecule 5mC is a limitation. In addition, while SMRT-based bisulfite sequencing allows sequencing lengths of up to ~2 kilobases (kb), it is dependent on bisulfite conversion.
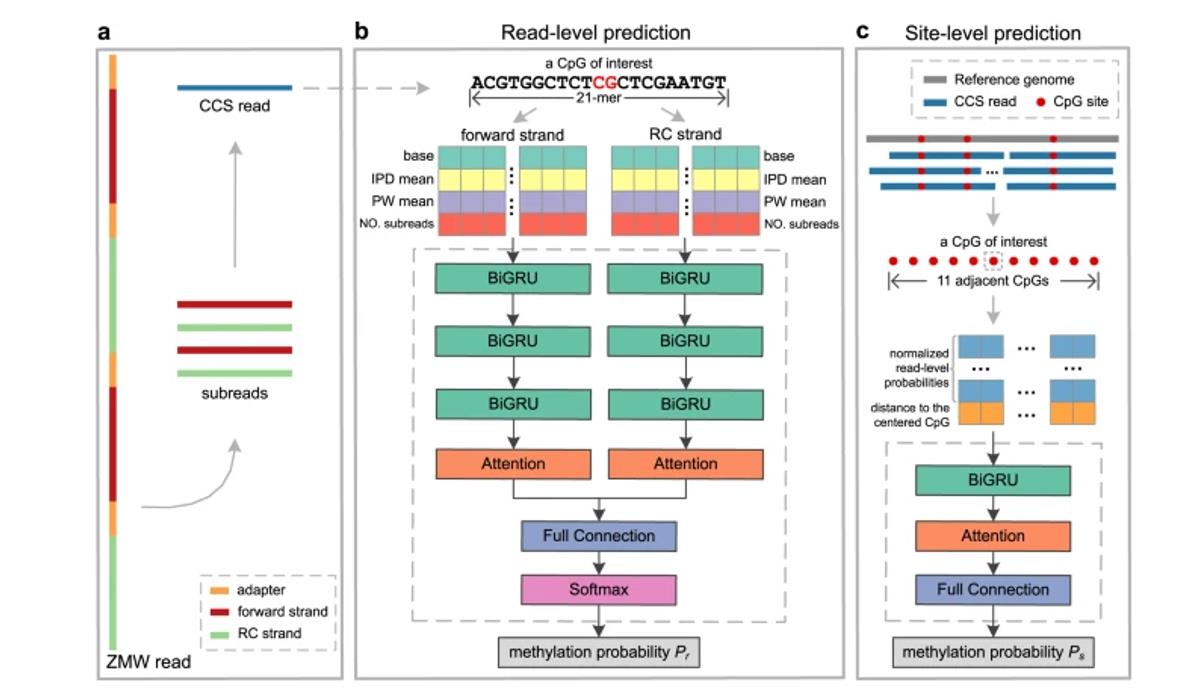 ccsmeth for 5mCpG detection using PacBio CCS reads. (Ni et al., 2023)
ccsmeth for 5mCpG detection using PacBio CCS reads. (Ni et al., 2023)
Nanopore Sequencing for 5mC Analysis
Nanopore sequencing technology detects DNA modifications by the difference in current intensity generated by nanopore reads of unmodified and modified bases. Specifically, the current pattern (also known as the "waveform curve") generated by the passage of a modified base through a pore is different from the current pattern generated by the passage of an unmodified base. After the nanopore reading bases are identified and compared, the differences can be determined by (1) statistical tests comparing the current patterns to a computer reference or patterns from unmodified control samples, and (2) pre-trained supervised learning models such as neural networks, machine learning models, and Hidden Markov Models (HMMs). Previous studies have shown that methods using pre-trained models can achieve high accuracy in DNA 5mC detection from human nanopore reads.
Limitations of Nanopore Sequencing:
The absence of amplification and prior enzymatic or chemical processing steps in nanopore sequencing supports the analysis of naturally modified DNA molecules. However, DNA methylation detection using nanopore sequencing poses a methodological challenge in terms of the ability to detect modifications of different CpGs in very close proximity to each other on DNA fragments, since it is assumed that all CpGs within a 10 bp region have the same methylation status. In order to detect non-canonical bases such as 5mC, trained computational tools such as Nanopolish, DeepSignal, Tombo and DeepMod are required.
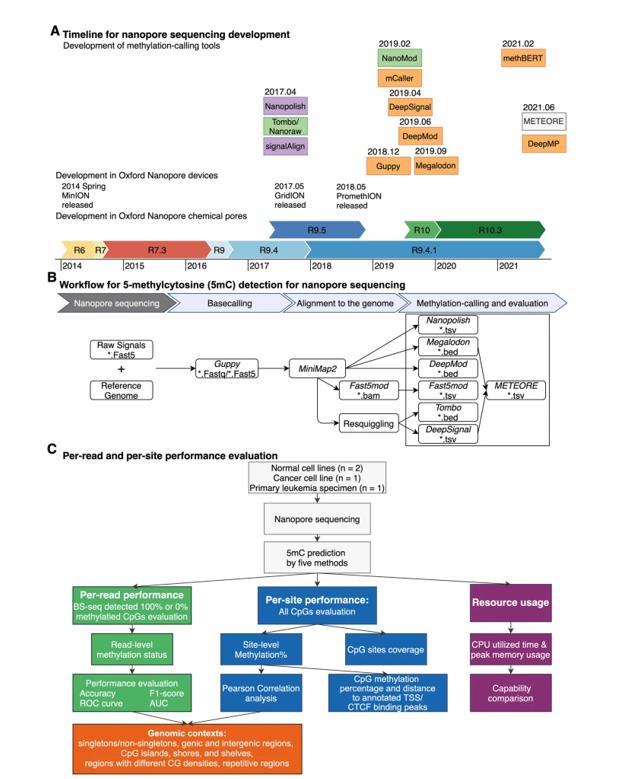 Technological development of methylation-calling tools and benchmark strategy of Oxford Nanopore Technologies (ONT). (Liu et al., 2021)
Technological development of methylation-calling tools and benchmark strategy of Oxford Nanopore Technologies (ONT). (Liu et al., 2021)
Comparison of PacBio and Nanopore Sequencing in 5mC Analysis
Depth and Breadth of Detection
Recent benchmarking has shown that Nanopore sequencing identifies more CpG sites associated with 5mC than PacBio's HiFi sequencing at the same sequencing depth. Specifically, Nanopolish enhanced the detection of methylation status by approximately 6% of CpG, which can be elusive for PacBio's approach. One implication of this observation is that Nanopolish may have a slight advantage for projects aimed at comprehensively characterizing genomic landscapes.
Performance Metrics
Precision, accuracy, recall, and F1 score are the most important metrics in any bioinformatics analysis. In the field of 5mC analysis, Nanopore demonstrates excellent performance on these metrics, making it more accurate and cost-effective for genome-wide DNA methylation detection.
Computational Requirements
While nanopore sequencing provides a wealth of data, it requires additional computational resources. Guppy's base identification process can be resource-intensive. PacBio's HiFi sequencing is less burdensome in this regard but compensates by requiring a higher sequencing depth.
In-depth Analysis and Applications
The strength of Nanopore is evident when it comes to detecting 5mC in biologically relevant genomic environments such as gene regions and repeat regions. This granularity makes Nanopore the first choice for in-depth epigenetic studies, providing a wealth of information for downstream applications.
References
- Liu, Yadong, et al. "Comparison of the Nanopore and PacBio sequencing technologies for DNA 5-methylcytosine detection." 2022 IEEE International Conference on Bioinformatics and Biomedicine (BIBM). IEEE, 2022.
- Ni, Peng, et al. "DNA 5-methylcytosine detection and methylation phasing using PacBio circular consensus sequencing." Nature Communications. 14.1 (2023): 4054.
- Liu, Yang, et al. "DNA methylation-calling tools for Oxford Nanopore sequencing: a survey and human epigenome-wide evaluation." Genome biology. 22.1 (2021): 1-33.
Related Services
PacBio SMRT Sequencing TechnologyOxford Nanopore Sequencing Technology
Long-Read Sequencing of DNA Methylation
For research purposes only, not intended for personal diagnosis, clinical testing, or health assessment