We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy


We are dedicated to providing outstanding customer service and being reachable at all times.
Nanopore Targeted Sequencing
In the dynamic realm of genomic exploration, addressing challenges head-on has led to groundbreaking advancements. Target sequencing emerges as a strategic beacon, illuminating regions of interest with unparalleled depth and precision. At CD Genomics, we seize this strategy and elevate it further through the integration of cutting-edge Nanopore Sequencing technology, revolutionizing the landscape of genomic analysis.
Based on this technology, we also provide Human and Animal/Plant Long Amplicon Sequencing services. If you have any specific questions about these sequencing services or if you'd like more detailed information about the technology, applications, or processes involved, feel free to ask!
Advantages of Nanopore Target Sequencing
- Enrichment of Inaccessible Regions: Enrich and sequence regions that are difficult or impossible to access using traditional sequencing technologies. This is particularly useful for analyzing genomic regions with complex structural variations, repetitive sequences, or high GC content.
- Different Enrichment Strategies: PCR, PCR-free enrichment and novel methods, such as CRISPR/Cas9. Learn more about adaptive sampling in PCR-free enrichment.
- No Read Length Restrictions: Characterize regions of interest without being constrained by maximum read lengths the same read length limitations as some other technologies.
- Wide Range of Target Sizes: Handle a broad range of target sizes, from short fragments (20 bp) to very long fragments (over 4 Mb), providing flexibility in experimental design.
- Comprehensive Data: Rich data enable the detection of single nucleotide variants (SNVs), structural variations (SVs), repeat expansions, and even modified bases within the targeted regions.
- On-Demand Sequencing: Tailored to specific applications, allowing you to sequence what you need, when you need it. This flexibility is particularly useful for various research and diagnostic purposes.
- Rapid Turnaround Times: The combination of rapid enrichment, library preparation methods, real-time sequencing, and analysis can lead to turnaround times of hours or less, providing quick access to results.
Our Service Workflow
Sample Requirements
High-quality nucleic acids: Genomic DNA ≥ 5 µg, concentration ≥ 20 ng/µL; Microbial DNA ≥ 500 ng, concentration ≥ 10 ng/µL
Since we offer amplicon preparation services, we also accept other samples, such as cell pellets, and other tissue samples.
We Can Help Customers Achieve
- SNVs Detection: Utilize Nanopore targeted sequencing for accurate single nucleotide variant (SNV) identification, crucial in disease genetics and evolution.
- Structural Variation Analysis: Detect deletions, insertions, inversions, and translocations with superior precision and resolution.
- Genomic Variant Phasing: Achieve detailed allele arrangement insights for better trait and disease understanding.
- Epigenetic Studies: Study DNA methylation and modifications in specific regions, revealing gene regulation insights.
- Gene Identification: Validate target genes comprehensively, aiding disease mutation and functional element discovery.
- Microbiome Analysis: Probe microbial genomic diversity for ecological and functional understanding.
- Pathogen Detection: Rapidly identify pathogens by targeting associated genes for effective disease management.
Case Studies
Case Study 1: Nanopore Cas9-Targeted Sequencing (nCATS) for Comprehensive Variant Detection
Existing methods have limitations such as loss of native modifications, short read lengths, high input requirements, low yield, and lengthy protocols. Addressing these limitations, the authors introduce Nanopore Cas9-Targeted Sequencing (nCATS) as a novel enrichment strategy that leverages Cas9-mediated targeted cleavage of chromosomal DNA to enable high sequencing depth and comprehensive variant detection.
This approach allows simultaneous assessment of various genetic features, including haplotype-resolved single-nucleotide variants, structural variations, and CpG methylation. The nCATS technique was applied to multiple samples, including cell lines, cell-line-derived xenografts, and primary human breast tissues (both normal and tumor/normal paired).
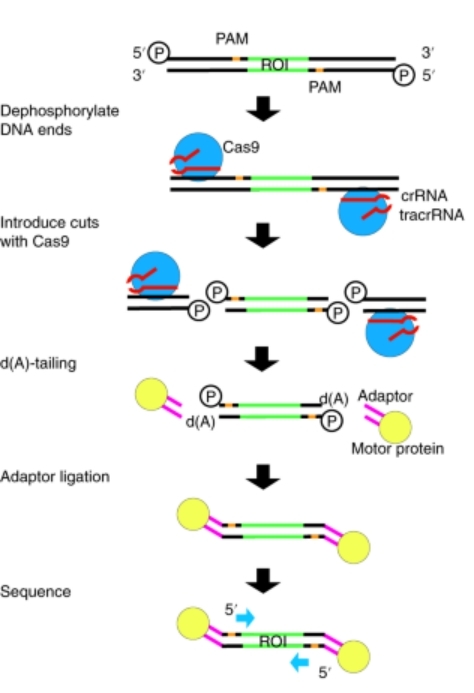
The results of the study demonstrated the robustness and effectiveness of the nCATS method. Median sequencing coverage reached 675× using the MinION flow cell and 34× using the smaller Flongle flow cell. Remarkably, nCATS required only approximately 3 μg of genomic DNA, which is notably lower than many existing methods. This efficient utilization of input DNA makes the technique more accessible and cost-effective.
The development of nCATS addresses the limitations of current sequencing methods by offering a powerful approach for comprehensive variant detection. The ability to simultaneously assess single-nucleotide variants, structural variations, and CpG methylation provides researchers and clinicians with a more complete view of genomic information.
Q&A
How should I decide whether to use Cas9 targeted sequencing, adaptive sampling, or PCR-based methods?
A: The choice between Cas9 targeted sequencing, adaptive sampling, and PCR-based methods depends on your specific research goals and sample characteristics. If you need to sequence challenging regions while retaining base modifications, Cas9 targeted sequencing and adaptive sampling are excellent choices. However, for low sample inputs or the detection of low-frequency variants, PCR may be essential to achieve the desired depth of coverage. Our expert team can help you make the right decision based on your project's requirements.
How does PCR contribute to achieving high depth of coverage?
A: PCR, or Polymerase Chain Reaction, is a widely used DNA amplification technique that can help increase the number of target DNA molecules in a sample before sequencing. By selectively replicating specific DNA regions, PCR can ensure that even low-frequency variants are present in sufficient quantities for a thorough analysis. This is particularly valuable when working with limited sample inputs or aiming to detect rare genetic variations.
Can I sequence multiple samples in multiplex without additional amplification steps?
A: Absolutely! Our Barcoding technology allows you to ligate a unique barcode to each amplified sample. Following barcoding, you can pool the samples and sequence them on a single flow cell. This multiplexing approach eliminates the need for further amplification, saving time and resources.
What nanopore sequencing platforms are available for targeted sequencing?
A: Our platforms cater to diverse needs. The portable MinION is laptop-operable, while the MinION Mk1C integrates a display and onboard computation for all-in-one sequencing and analysis. These are excellent for single targets or smaller panels. For more information, you can refer to our article Nanopore Sequencing 101: Choosing the Right Sequencing Device.
Reference
- Gilpatrick, Timothy, et al. "Targeted nanopore sequencing with Cas9-guided adapter ligation." Nature biotechnology 38.4 (2020): 433-438.
For research purposes only, not intended for personal diagnosis, clinical testing, or health assessment