

We are dedicated to providing outstanding customer service and being reachable at all times.
Long-Read Metagenomics Sequencing
Metagenomics serves as a powerful approach to improve our biological insight into the microbes living in underexplored environments, involving the identification of unknown, uncultured microorganisms as well as the discovery of unknown functional genes. Our metagenomics sequencing service is based on advanced PacBio and Nanopore sequencing platforms, which combine the sample characteristics and data output to select the optimal assembly effect and fully explore the microbial communities and functional genes in environmental samples.
Challenges for Metagenome-Assembled Genomes (MAGs)
Metagenomics typically includes genome-wide assembly, taxonomic analysis, and functional profiling, which allows researchers to gain insight into microbial communities. This method aims to sequence the entire genomic content of microbial samples, thus providing greater genomic information than targeted approaches. Metagenomics sequencing allows not only the accurate recovery of strain-level information but also the detection and identification of specific genes, such as antimicrobial resistance (AMR) genes or other virulence elements. In addition, it helps to identify virus communities missed by targeted sequencing methods. The next-generation sequencing (NGS) technologies have allowed enormous datasets that can be used for the assembly of mock genomes called metagenome-assembled genomes (MAGs). With high-throughput low-error short-read sequencing, such as Illumina, MAGs have led to a real revolution in the area of microbiology during the last 10 years. However, assembly driven metagenomics has disadvantages, including,
- Low recovery of high micro-diversity microbes.
- Low recovery of the resilient genome.
- Uncertainty due to potential chimera production.
Introduction of Long-read Metagenomics Sequencing
Long-read metagenomics sequencing, including PacBio and Oxford Nanopore Sequencing Technology (ONT) platform, is not limited by NGS short-read length. Long-read sequencing technologies far surpass NGS technologies in terms of assembly completeness, species annotation accuracy, gene completeness, and analysis of the association between resistance genes and species affiliation, thus better facilitating the elucidation of the mechanisms by which microbial communities function in ecosystems.
Based on PacBio's long-read length, it can easily cover intergenic regions or gene-specific regions, giving more realistic feedback on the complex composition of the colony. While the ONT Nanopore sequencing platform has become a new hotspot for metagenomics sequencing analysis with its ultra-long read length and lower sequencing cost.
Sample Requirements
- Sample Type: Genomic DNA/cDNA, OD260/280=1.8~2.0, no degradation and no contamination.
- DNA amount:
-PacBio Sequel ll platform: ≥ 8μg,
-Nanopore PromethION platform: ≥ 8μg.
Analysis Workflow
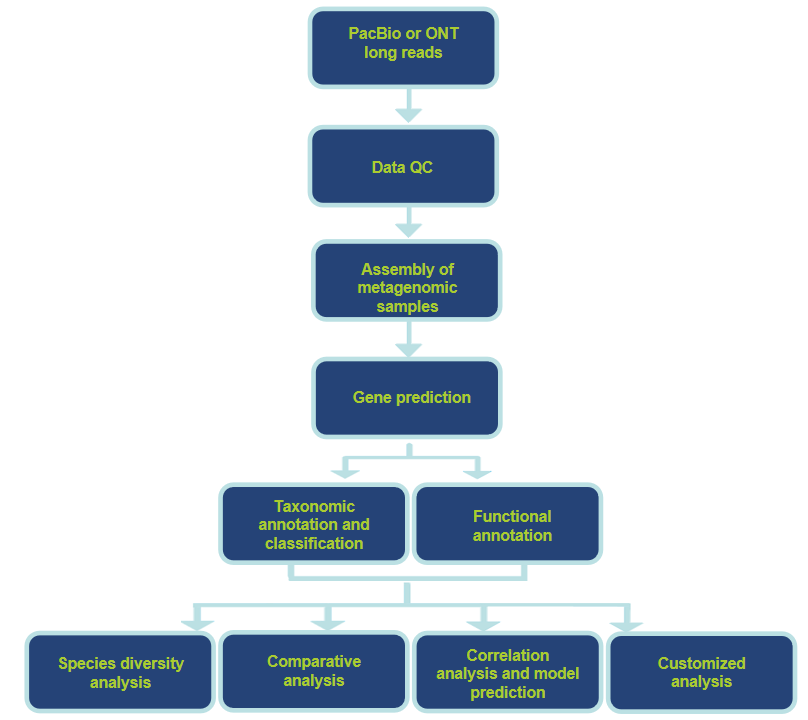
Analysis Contents
| Standard Analysis | Advanced Analysis |
| Sequencing data quality assessment | Species diversity analysis |
| Metagenomic assembly | Comparative analysis |
| Gene prediction | Similarity analysis |
| Taxonomic and functional annotation | Correlation analysis and model prediction |
| Genome reconstruction | Customized analysis |
Long-read metagenomic assembly not only improves the continuity of short-read metagenome-assembly, but also improves strain resolution, sequencing of new plasmids and viruses, and enhances the ability to identify horizontal gene transfer. To perform long-read metagenomics sequencing, researchers and professionals choose CD Genomics. Based on several experts in the field and many years of experience, our services provide highly reliable, turnaround time and cost-effective metagenomics sequencing and analysis. Please feel free to contact us. We've got everything covered for your needs and are ready to assist.
For research purposes only, not intended for personal diagnosis, clinical testing, or health assessment
