We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy

Non-coding RNAs (ncRNAs) are RNA molecules that do not encode proteins but regulate gene expression through various mechanisms. In recent years, ncRNAs have emerged as critical regulators of epigenetic processes, drawing increasing attention for their roles in both health and disease. This review explores the epigenetic functions of ncRNAs and the role of bioinformatics in elucidating their complex regulatory networks.
Non-coding RNAs encompass a diverse group of molecules, including microRNAs (miRNAs), small interfering RNAs (siRNAs), PIWI-interacting RNAs (piRNAs), and long non-coding RNAs (lncRNAs). These ncRNAs modulate gene expression at transcriptional and post-transcriptional levels, influencing chromatin dynamics, DNA methylation, histone modifications, and RNA stability. The primary mechanisms through which ncRNAs exert epigenetic control are summarized below:
miRNAs: Small (~22 nucleotides) regulatory RNAs that bind to complementary sequences on target mRNAs, leading to their degradation or translational repression. miRNAs play significant roles in gene silencing and are implicated in various diseases, including cancer.
siRNAs: Double-stranded RNA molecules that guide the RNA-induced silencing complex (RISC) to complementary mRNA targets, facilitating mRNA cleavage and gene silencing. siRNAs are primarily involved in antiviral defense and transposon silencing.
piRNAs: A distinct class of small RNAs that interact with PIWI proteins, primarily functioning in germline cells to silence transposable elements and maintain genome integrity.
lncRNAs: RNA molecules longer than 200 nucleotides that regulate gene expression through diverse mechanisms, including chromatin remodeling, transcriptional interference, and acting as molecular scaffolds. lncRNAs are involved in a wide range of cellular processes, from X-chromosome inactivation to the regulation of developmental pathways.
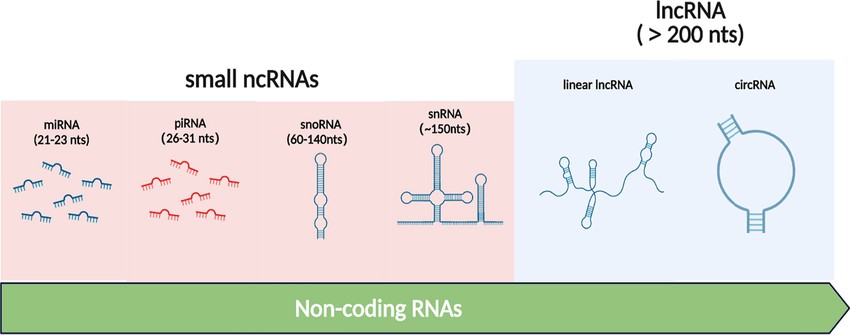 Figure 1. Types of Non-coding RNAs (Jie-yu Sun et al,. 2021)
Figure 1. Types of Non-coding RNAs (Jie-yu Sun et al,. 2021)
ncRNA molecules regulate gene expression through interactions with DNA, RNA, or proteins, utilizing various mechanisms to control cellular growth, differentiation, and function. Recent studies have demonstrated that ncRNAs also modulate epigenetic marks, such as DNA methylation and histone modifications, thereby influencing gene expression.
ncRNAs function through diverse pathways, impacting gene expression at multiple levels:
Interaction with DNA: ncRNAs can guide chromatin-modifying complexes to specific genomic loci, influencing chromatin state and accessibility. For instance, lncRNAs can recruit DNA methyltransferases, leading to targeted DNA methylation, which suppresses gene transcription.
RNA Interference: Small ncRNAs, such as miRNAs and siRNAs, interact with complementary RNA sequences to mediate post-transcriptional gene silencing. These molecules are integral to the RNA-induced silencing complex (RISC), where they guide the complex to target mRNAs, resulting in degradation or translational repression.
Histone Modification: ncRNAs influence histone modifications, such as acetylation, methylation, and phosphorylation, which are key determinants of chromatin structure and gene expression. For example, certain lncRNAs serve as scaffolds for chromatin-modifying enzymes, directing them to specific histone residues to alter chromatin conformation and regulate transcriptional activity.
Modulation of Epigenetic Landscapes: Emerging evidence suggests that ncRNAs are critical in shaping the epigenetic landscape by directly influencing DNA methylation patterns and histone modifications. This regulation can be gene-specific or genome-wide, contributing to the dynamic control of gene expression in response to cellular signals and environmental changes.
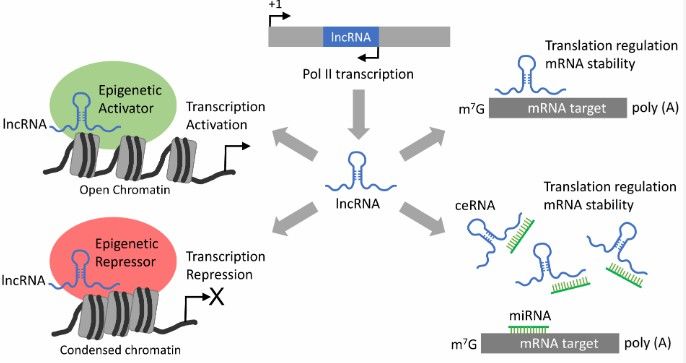 Figure 2. LncRNA-mediated gene regulation mechanisms. (Subhasree Kumar et al,. 2022)
Figure 2. LncRNA-mediated gene regulation mechanisms. (Subhasree Kumar et al,. 2022)
Epigenetic research on ncRNAs represents a rapidly evolving field with substantial potential for elucidating gene regulation and disease pathogenesis. The study of ncRNA-mediated epigenetic regulation is facilitated by various advanced techniques that allow for the exploration of ncRNA expression patterns, interactions with epigenetic marks, and functional roles in different cellular contexts. The following methodologies are commonly employed in the study of ncRNA epigenetics:
RNA sequencing is a powerful technique employed to determine differential expression patterns of ncRNAs. This method enables the identification of ncRNAs that exhibit variable expression across different cells or tissues, providing insights into the roles of ncRNAs in epigenetic regulation. By analyzing the expression profiles of ncRNAs, researchers can elucidate their contributions to gene expression modulation in health and disease.
ChIP-seq is a widely utilized technique for identifying the interactions between ncRNAs and epigenetic marks. This approach aids in determining how ncRNAs regulate specific gene expression by examining their interactions with chromatin-associated proteins and epigenetic modifications such as histone marks. ChIP-seq provides valuable data on the regulatory networks mediated by ncRNAs and their influence on chromatin dynamics.
Service you may intersted in
Immunoprecipitation techniques, including RNA immunoprecipitation (RIP) and crosslinking immunoprecipitation (CLIP), are employed to identify interactions between ncRNAs and proteins. By isolating complexes of ncRNAs with specific proteins, these methods provide insights into the mechanistic roles of ncRNAs in modulating epigenetic marks. The identification of protein partners of ncRNAs enhances the understanding of their functional roles in gene regulation.
In situ hybridization is a pivotal technique used to study the spatial expression patterns of ncRNAs within cells and tissues. This method enables the localization of ncRNAs at the subcellular level, revealing their distribution and providing clues about their functional roles in epigenetic regulation. The precise mapping of ncRNA expression helps elucidate their involvement in specific cellular processes.
CRISPR-Cas9 genome editing technology is instrumental in studying the functional roles of ncRNAs in epigenetic regulation by enabling targeted knockout or overexpression of specific ncRNAs. By manipulating ncRNA expression, researchers can investigate the effects on gene expression and epigenetic marks, thereby uncovering the regulatory mechanisms governed by ncRNAs.
Single-cell sequencing techniques allow for the investigation of ncRNA expression at the single-cell level, revealing expression heterogeneity among different cell types. This approach provides a high-resolution view of ncRNA function in epigenetic regulation, highlighting cell-specific roles and regulatory patterns that are often obscured in bulk analysis. The ability to analyze individual cells enhances the understanding of ncRNA-mediated gene regulation in complex tissues.
The investigation of epigenetic mechanisms involving ncRNAs requires a systematic approach to address relevant biological questions, define the study population, collect and analyze data, validate findings, and explore potential clinical applications. The following steps outline a general framework commonly employed in the study of ncRNA-mediated epigenetic regulation:
The initial step involves the identification of the specific research question. This may include investigating the role of ncRNAs in a particular disease or pathological condition, or identifying novel ncRNAs implicated in epigenetic regulation. Defining the research question provides a clear focus and guides the subsequent experimental design.
The definition of the study population is critical and should align with the research question. This may involve selecting patients with a specific disease, healthy controls, or other relevant groups. The choice of study population determines the scope of the investigation and influences data interpretation.
Genomic data are typically collected from public databases or generated using advanced techniques such as RNA-seq and ChIP-seq. These technologies facilitate the identification of differentially expressed ncRNAs and associated epigenetic marks. Bioinformatics tools are then employed to analyze the data, elucidating the relationships between ncRNAs and epigenetic modifications.
Upon identifying candidate ncRNAs and epigenetic marks, it is essential to validate the results using additional experimental approaches, such as quantitative polymerase chain reaction (qPCR) or in situ hybridization. These validation steps confirm the initial findings and ensure the reliability of the identified ncRNA-epigenetic interactions.
Once candidate ncRNAs have been validated, functional analysis is conducted to elucidate their roles in epigenetic regulation. This may involve downstream target analysis, such as knockdown or overexpression experiments, to assess the impact of ncRNAs on gene expression and epigenetic modifications. Functional assays provide insights into the mechanistic pathways through which ncRNAs influence cellular processes.
If the ncRNAs are found to play significant roles in epigenetic regulation, further research may explore their potential as biomarkers or therapeutic targets. Clinical applications could involve evaluating ncRNAs as diagnostic tools, prognostic indicators, or therapeutic modulators, thereby expanding their relevance in personalized medicine.
Bioinformatics tools are increasingly employed to analyze large-scale genomic data and to identify novel ncRNAs involved in epigenetic regulation. Researchers utilize these tools to analyze RNA-seq data, ChIP-seq data, and other high-throughput genomic datasets to identify differentially expressed ncRNAs or those associated with specific epigenetic modifications.
Bioinformatics approaches have been instrumental in elucidating the roles of ncRNA epigenetics in cancer development and progression. Through the use of computational tools, dysregulated ncRNAs have been identified across various cancer types, demonstrating their involvement in modulating epigenetic marks to control gene expression. This research underscores the potential of bioinformatics to reveal critical ncRNA functions and their impacts on epigenetic landscapes in oncogenesis.
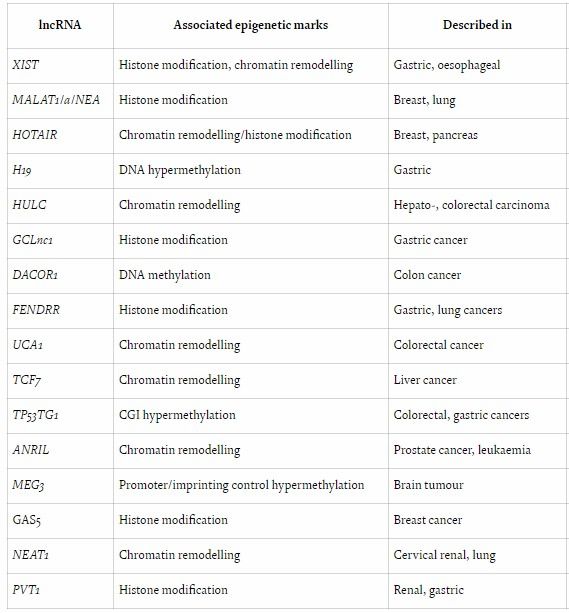
The following provides an overview of several commonly used R packages and their applications in the study of ncRNA-epigenetic interactions:
ChIPseeker is an R package designed for the annotation and analysis of ChIP-Seq data. It provides gene annotation, Gene Ontology (GO) annotation, and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotation, enabling the study of interactions between ncRNAs and chromatin modifications. ChIPseeker is particularly valuable for investigating how ncRNAs influence chromatin structure and gene regulatory regions.
DiffBind is an R package used for the differential analysis of ChIP-Seq data. It evaluates the effects of histone modifications, DNA methylation, and transcription factor binding on gene expression. This package is instrumental in studying the interactions between ncRNAs and various epigenetic modifications, allowing researchers to assess how changes in chromatin accessibility impact ncRNA function.
methylKit is an R package for analyzing methylation data, specifically designed to evaluate changes in DNA methylation patterns and distribution. It can be employed to explore the interactions between ncRNAs and DNA methylation, providing insights into how ncRNAs may modulate methylation landscapes to influence gene expression.
edgeR is an R package for differential expression analysis of RNA-Seq data. It evaluates changes in gene expression and is widely used to study the interactions between ncRNAs and gene expression levels. By analyzing differential expression, edgeR aids in identifying ncRNAs that may regulate or be regulated by transcriptional activity.
clusterProfiler is an R package used for functional enrichment analysis, including GO, KEGG, and Reactome pathway enrichment. It is utilized to study the interactions between ncRNAs and functional pathways, providing insights into the broader biological roles of ncRNAs in cellular processes.
DESeq2 is an R package for differential expression analysis of RNA-Seq data. In addition to evaluating gene expression changes, it offers gene annotation and functional enrichment analysis. DESeq2 is essential for studying the interactions between ncRNAs and gene expression, enabling researchers to identify regulatory networks involving ncRNAs.
limma is an R package designed for the differential expression analysis of microarray and RNA-Seq data. It assesses changes in gene expression and provides gene annotation and functional enrichment analysis. This package is used to explore the interactions between ncRNAs and gene expression, offering a robust framework for understanding ncRNA regulatory functions.
EpiDISH is an R package used for sample classification and grouping based on DNA methylation data. It can be employed to investigate the interactions between ncRNAs and sample classification, helping to elucidate the role of ncRNAs in disease subtyping and epigenetic heterogeneity.
MethPipe is an R package for analyzing methylation data, including the evaluation of DNA methylation changes and distribution. It provides gene annotation and functional enrichment analysis, making it suitable for studying the interactions between ncRNAs and DNA methylation, particularly in the context of epigenetic regulation.
biomaRt is an R package for gene annotation and data retrieval from multiple databases. It is used to obtain gene annotation information relevant to ncRNAs and their interactions, offering a comprehensive resource for researchers studying ncRNA function and regulation.
The utilization of ncRNAs as biomarkers for disease diagnosis and prognosis represents a promising area of research. By analyzing ncRNA expression patterns in patient samples, researchers can identify novel biomarkers for early disease detection and monitoring of disease progression.
Bioinformatics tools play a critical role in this field by facilitating the identification of ncRNAs associated with specific disease states. These tools enable the comprehensive analysis of large datasets to uncover ncRNAs that correlate with disease phenotypes, thereby aiding in the development of predictive models for disease progression. Such models are instrumental in advancing precision medicine, providing valuable insights into the molecular underpinnings of disease and offering potential pathways for targeted therapeutic interventions.
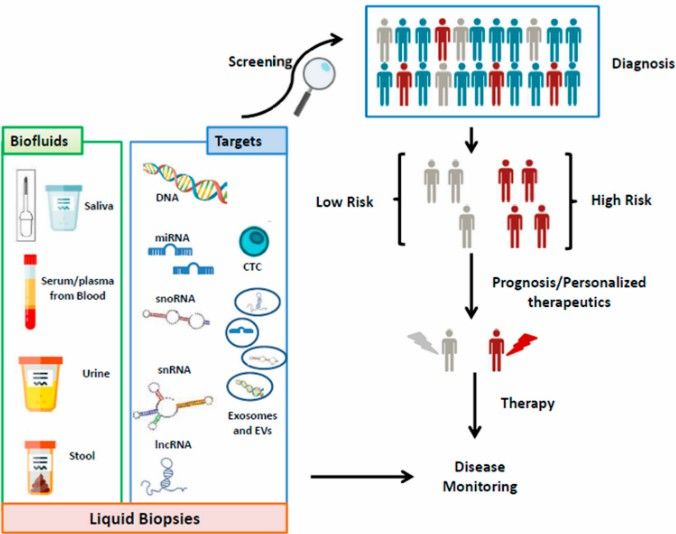 Figure 3. An effective management of cancer diagnosis screening by using body fluids and molecular biomarkers (Barbara Pardini et al,.2019)
Figure 3. An effective management of cancer diagnosis screening by using body fluids and molecular biomarkers (Barbara Pardini et al,.2019)
With the rapid advancements in epigenetics, the roles of ncRNAs in cellular biology have garnered significant attention. Accumulating evidence suggests that ncRNAs function through various mechanisms, including the regulation of gene expression, modulation of cellular differentiation, and participation in disease pathogenesis. Consequently, exploring the epigenetic landscapes of ncRNAs has emerged as a critical area of contemporary research.
The investigation of ncRNA epigenetics offers valuable insights for disease diagnosis and therapeutic interventions. For instance, several studies have demonstrated that ncRNAs play pivotal roles in the initiation and progression of tumors, highlighting the potential of ncRNA epigenetic landscapes as novel diagnostic and therapeutic targets in oncology. Furthermore, the study of ncRNA epigenetics can elucidate the mechanisms of cellular differentiation, providing a deeper understanding of ncRNAs in developmental biology, immune regulation, and growth processes.
As epigenetic technologies continue to evolve, the research on ncRNA epigenetic landscapes is poised for substantial growth. The advent of single-cell sequencing technologies enables high-resolution analysis of ncRNA expression and regulatory dynamics at the individual cell level. Additionally, emerging epigenetic techniques and analytical methods promise to enhance the precision and depth of ncRNA epigenetic landscape studies, offering new avenues for scientific exploration.
References
Terms & Conditions Privacy Policy Copyright © CD Genomics. All rights reserved.