Introduction to Mouse CpG Island Microarray Service
DNA methylation is a central regulatory mechanism in epigenetics, exerting a profound influence on gene expression, cell fate, and the pathogenesis of various diseases. Investigating the methylation status of CpG islands in the mouse genome is pivotal for deciphering gene regulation, developmental processes, and disease mechanisms. To accurately quantify changes in CpG island methylation, microarray technology has emerged as an efficient tool.
Research into mouse epigenetics employs diverse technologies, with CD Genomics offering comprehensive solutions such as the Agilent Mouse CpG Island Microarray and Illumina Infinium Mouse Methylation BeadChip services. These services encompass a thorough coverage of CpG islands and methylation sites across the mouse genome, thereby facilitating advanced epigenetic research.
The Agilent Mouse CpG Island Microarray represents a state-of-the-art technology in this domain, utilizing Agilent's 60-mer SurePrint technology to cover 16,030 CpG islands within the mouse genome. This microarray is designed with 97,652 probes, enabling precise measurement of DNA methylation states. Each array is configured as 2x105K, comprising 105,000 probes per array, making it suitable for high-throughput and accurate whole-genome methylation analysis.
 Agilent Microarray.
Agilent Microarray.
Source: Agilent Technologies
CD Genomics provides mouse CpG island microarray services based on this technology, delivering high-precision and high-sensitivity methylation data to researchers. Through this service, researchers can delve deeply into gene expression regulation, unravel disease mechanisms, and explore epigenetic processes. Whether for cancer research, developmental biology, or other biological fields, CD Genomics' mouse CpG island microarray services offer robust data support.
Principle of Agilent Microarray Detection
The Agilent Mouse CpG Island Microarray technology employs a high-density probe design to conduct a comprehensive and systematic analysis of the methylation status of CpG islands within the mouse genome. The underlying principle is as follows:
Genomic DNA is digested using two restriction enzymes, Alu I and Rsa I. The Alu I enzyme recognizes and cleaves at the "AGCT" sequence. In cases where the genomic site is a "CGCT" homozygote, it remains uncut, thereby preserving intact long DNA strands. These intact strands exhibit strong binding affinity to the probes, resulting in high-intensity fluorescence signals during laser scanning. Conversely, if the site is a heterozygote (CGCT/AGCT), the "AGCT" allele is cleaved while the "CGCT" allele remains intact. During subsequent hybridization, only the intact long strand hybridizes efficiently with the probes, while the shortened cleaved fragments exhibit reduced probe affinity and are washed away during the elution process. Consequently, only the uncut long strand from the two alleles remains on the probe, yielding intermediate fluorescence intensity.
In the case where the genomic site is an "AGCT" homozygote, the enzymes completely cleave the DNA, which, upon hybridization, is entirely washed away. As a result, the probe exhibits either no fluorescence or very low fluorescence intensity.
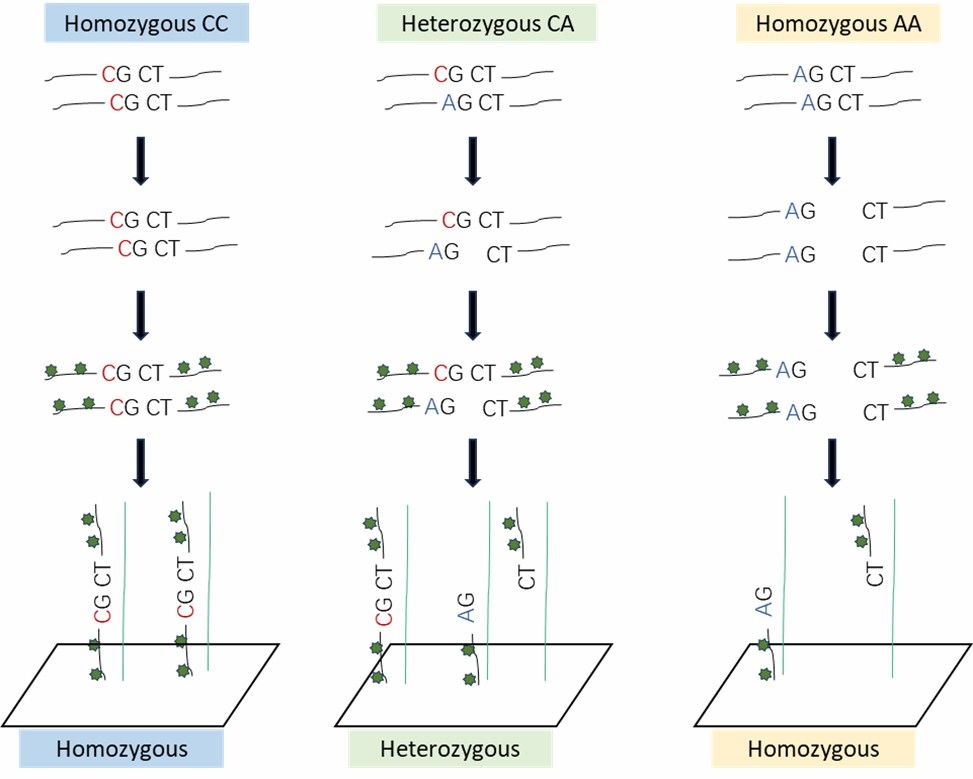 Agilent Chip Detection Principle.
Agilent Chip Detection Principle.
Technical Advantages
- High Throughput and Comprehensive Coverage: The Agilent Mouse CpG Island Microarray encompasses 16,030 CpG islands, delivering extensive genomic data suitable for large-scale methylation research. The capability to simultaneously analyze multiple CpG islands on each chip significantly enhances experimental efficiency.
- High Precision and Sensitivity: Leveraging 60-mer SurePrint technology, the probe design of the microarray is exceedingly precise, capable of detecting methylation information even in samples with very low abundance. This ensures measurement results are characterized by high precision and sensitivity.
- Reliable Experimental Platform: Chips manufactured using Agilent SurePrint technology exhibit exceptional stability and reproducibility, guaranteeing consistent results across different experiments and batches.
- Versatile Applications: This technology is not only applicable to cancer research but also extends to developmental biology, genomic imprinting, and epigenetic studies, among other fields. It aids researchers in gaining deeper insights into the regulation of gene expression across various biological processes.
Workflow of the Mouse CpG Island Microarray Service
CD Genomics offers an extensive Agilent Mouse CpG Island Microarray technology, covering the complete workflow from sample preparation to data analysis, thus ensuring accurate and efficient measurement of methylation patterns. The main steps in this process include:
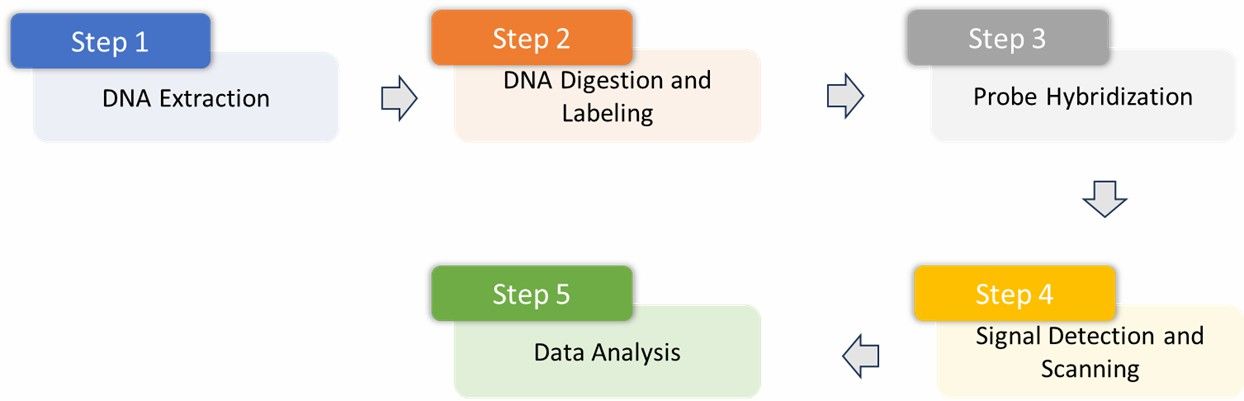 Agilent Chip Detection Principle.
Agilent Chip Detection Principle.
1. Sample Preparation and DNA Extraction: Genomic DNA of high quality is meticulously extracted from mouse tissues, with a focus on maintaining DNA integrity and purity. This is crucial for ensuring the reliability and validity of the experimental results.
2. DNA Digestion and Labeling: The extracted DNA is subjected to digestion using restriction enzymes, such as Alu I and Rsa I. Following digestion, the DNA is labeled with fluorescent tags to enable subsequent detection steps.
3. Probe Hybridization: The fluorescently labeled DNA is hybridized with the probes on the Agilent Mouse CpG Island Microarray. These probes are specifically engineered to target CpG island regions within the mouse genome, where the methylation status significantly influences the intensity of hybridization.
4. Signal Detection and Scanning: The microarray is scanned to detect fluorescent signals. Typically, methylated CpG islands are associated with weaker signals, whereas unmethylated regions exhibit stronger fluorescence, facilitating differential analysis.
5. Data Analysis: Advanced bioinformatics tools are used to analyze the detected data, allowing for the identification of methylation changes. The analysis culminates in a detailed and comprehensive report, highlighting significant epigenetic modifications.
Sample Requirements
- Sample Type: Acceptable samples include genomic DNA derived from mouse tissues, blood, cells, or other sources.
- Sample Quantity: A minimum of 10 μg of high-quality genomic DNA is required.
- Sample Purity: The A260/A280 ratio must be greater than 1.8 to ensure the DNA sample meets the purity standards necessary for the experiment.
Data Analysis and Delivery
Upon data generation, CD Genomics conducts thorough data analysis and provides clients with the following:
- Methylation Level Report: Detailed methylation status for each CpG island is provided, quantifying methylation levels and analyzing changes under different experimental conditions.
- Differential Methylation Analysis: Analysis of methylation differences between groups is performed to reveal potential biological disparities.
- Gene Function and Pathway Analysis: Methylated genes are functionally annotated using existing gene function databases, along with relevant biological pathway analyses.
- Comprehensive Data Report: The report includes experimental design, data analysis workflows, statistical graphs, and interpretations, assisting researchers in comprehending the experimental outcomes fully and offering plausible biological explanations.
Applications of Mouse CpG Island Microarray
The Mouse CpG Island Microarray Service provides a robust platform for examining DNA methylation patterns within CpG islands across the mouse genome. This tool is extensively utilized in fields such as epigenetics, disease research, developmental biology, and drug discovery. Through the analysis of CpG island methylation, researchers can acquire valuable insights into gene regulation, the mechanisms underlying diseases, environmental impacts, and the effects of genetic modifications.
Advantages of CD Genomics' Mouse CpG Island Microarray Service
- Leading Technology: Utilizing Agilent Mouse CpG Island Microarray technology, which encompasses 16,030 CpG islands, this service provides precise genomic methylation analysis.
- Extensive Expertise: CD Genomics has accumulated substantial experience in biotechnology services, particularly in epigenetics research and methylation analysis, offering clients high-quality technical support.
- Customized Service: Tailored to meet the specific needs of clients, CD Genomics provides flexible experimental design and personalized data analysis options, ensuring the most suitable results for each research project.
- Comprehensive Data Support: The reports provided by CD Genomics extend beyond methylation data, including thorough differential analysis, gene function, and biological pathway analysis, enabling clients to fully understand their experimental data.
Conclusion
While technologies like the Illumina Infinium Mouse Methylation BeadChip offer broad coverage and high-throughput capabilities at the whole-genome level, the Agilent Mouse CpG Island Microarray stands out for its precise measurement and high-density probe design focused specifically on CpG island regions. This makes it a powerful tool for investigating methylation status in the gene regulatory regions of the mouse genome. CD Genomics provides support for this advanced technology, aiding researchers in the thorough exploration of gene expression, epigenetics, and disease mechanisms.
 Description and technical and biological validation of the CpG sites mouse DNA methylation microarray. (Garcia-Prieto, Carlos A., et al. 2022)
Description and technical and biological validation of the CpG sites mouse DNA methylation microarray. (Garcia-Prieto, Carlos A., et al. 2022)
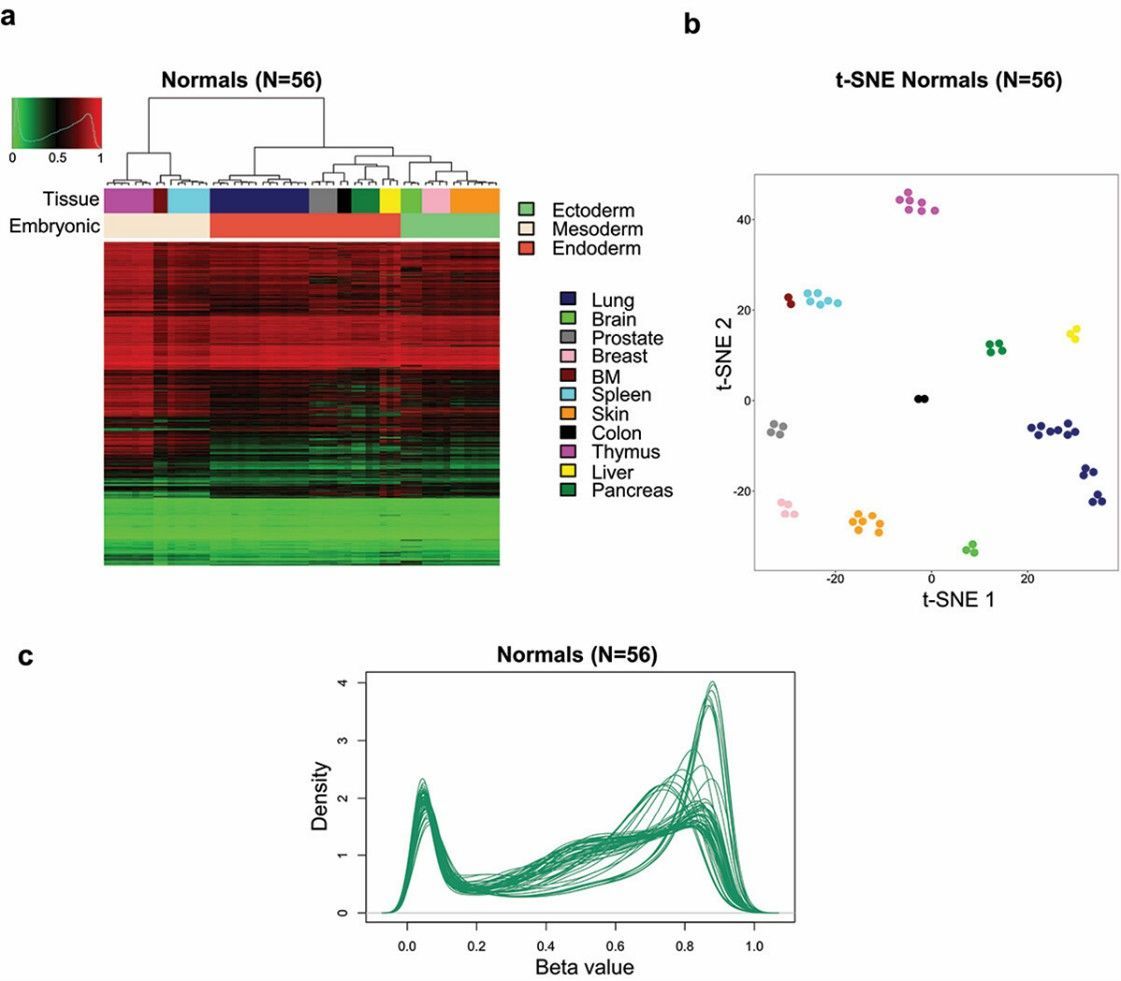 DNA methylation atlas for mouse normal tissues. (Garcia-Prieto, Carlos A., et al. 2022)
DNA methylation atlas for mouse normal tissues. (Garcia-Prieto, Carlos A., et al. 2022)
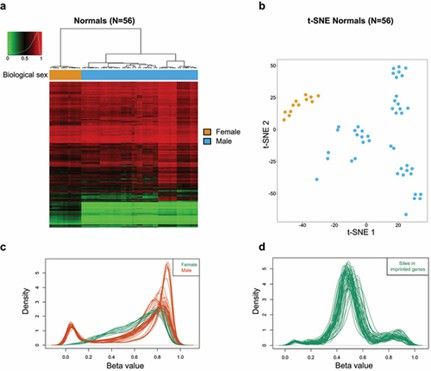 Mouse DNA methylation mapping according to biological sex, X-chromosome, and imprinted CpG sites. (Garcia-Prieto, Carlos A., et al. 2022)
Mouse DNA methylation mapping according to biological sex, X-chromosome, and imprinted CpG sites. (Garcia-Prieto, Carlos A., et al. 2022)
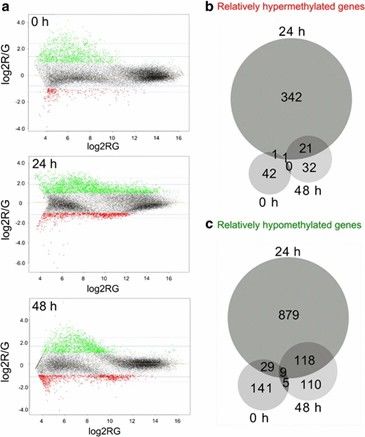 Significantly differentially methylated genes upon butyrate treatment. (Wippermann, A., et al., 2014)
Significantly differentially methylated genes upon butyrate treatment. (Wippermann, A., et al., 2014)
 Gene ontology analysis of genes affected by butyrate-mediated differential methylation of associated CpG islands. (Wippermann, A., et al., 2014)
Gene ontology analysis of genes affected by butyrate-mediated differential methylation of associated CpG islands. (Wippermann, A., et al., 2014)
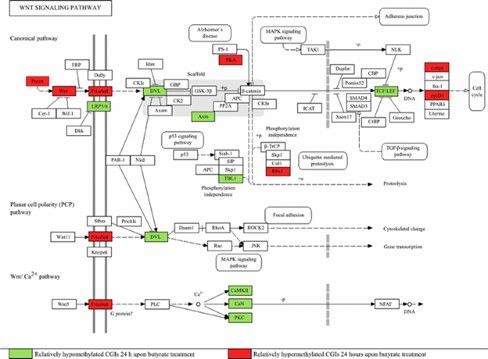 Differential methylation of CGIs associated with genes involved in the Wnt signal transduction system 24 h upon butyrate addition. (Wippermann, A., et al., 2014)
Differential methylation of CGIs associated with genes involved in the Wnt signal transduction system 24 h upon butyrate addition. (Wippermann, A., et al., 2014)
References
- Garcia-Prieto, Carlos A., et al. "Validation of a DNA methylation microarray for 285,000 CpG sites in the mouse genome." Epigenetics 17.12 (2022): 1677-1685. https://doi.org/10.1080/15592294.2022.2053816
- Wippermann, A., Klausing, S., Rupp, O. et al. Establishment of a CpG island microarray for analyses of genome-wide DNA methylation in Chinese hamster ovary cells. Appl Microbiol Biotechnol 98, 579–589 (2014). https://doi.org/10.1007/s00253-013-5282-2
