We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy

The CRISPR/Cas9 gene-editing system has emerged as a revolutionary tool in the field of genome editing, demonstrating exceptional efficiency in targeted DNA cleavage across various organisms, including humans, mice, and plants. However, a notable limitation of this system is its propensity for unintended binding of the Cas9-sgRNA complex to non-target regions, leading to off-target cleavage events. These off-target effects have been documented in numerous studies, and they pose a significant risk by potentially inducing undesirable consequences, thereby limiting the broader application of CRISPR/Cas9 in clinical settings.
Given these challenges, designing sgRNAs that exhibit both high on-target efficiency and minimal off-target activity has become an urgent research priority. Additionally, the accurate quantification of off-target effects is critical to enhancing the safety and efficacy of the CRISPR/Cas9 system. Previously, tools such as Cas-Offinder have been introduced, which provide an initial identification of potential off-target sites. However, these tools are limited in their ability to precisely evaluate the likelihood of off-target activity occurring at these sites.
Addressing this limitation, researchers led by Lin et al. from the University of Hong Kong have leveraged a recurrent convolutional neural network (RCNN) algorithm to develop an innovative tool called CRISPR-Net. This tool provides accurate quantification of off-target effects caused by mismatches, insertions, or deletions (Indels) between the sgRNA and target DNA during the CRISPR/Cas9 editing process. CRISPR-Net offers a more comprehensive and refined evaluation of off-target risk, equipping researchers with enhanced capabilities for assessing the safety of CRISPR-mediated genome editing.
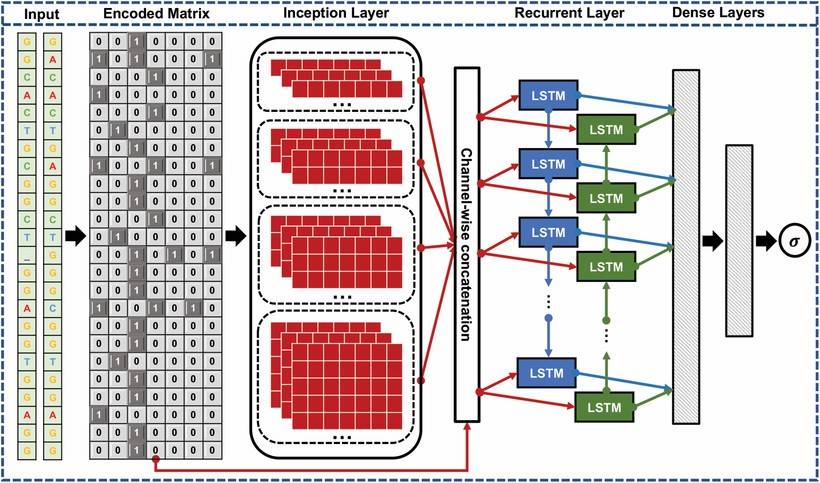 Figure 1 illustrates the architecture of CRISPR-Net. The user inputs a pair of sequences: the on-target sequence (XON) and the off-target sequence (XOFF). These sequences are encoded into binary matrices, which are then fed into the Inception layer of the convolutional neural network (CNN). The initial layer of the network contains 10 filters of varying sizes. The output from the Inception layer is integrated with the encoded matrices through feature fusion, and subsequently passed into the convolutional layer. The output from the convolutional layer is then dimensionally reduced and transmitted to two non-linear dense layers, consisting of 80 and 20 neurons, respectively. Specifically, the output neurons utilize the sigmoid activation function, while the remaining layers employ the ReLU activation function.
Figure 1 illustrates the architecture of CRISPR-Net. The user inputs a pair of sequences: the on-target sequence (XON) and the off-target sequence (XOFF). These sequences are encoded into binary matrices, which are then fed into the Inception layer of the convolutional neural network (CNN). The initial layer of the network contains 10 filters of varying sizes. The output from the Inception layer is integrated with the encoded matrices through feature fusion, and subsequently passed into the convolutional layer. The output from the convolutional layer is then dimensionally reduced and transmitted to two non-linear dense layers, consisting of 80 and 20 neurons, respectively. Specifically, the output neurons utilize the sigmoid activation function, while the remaining layers employ the ReLU activation function.
You may interested in
CD Genomics offers a range of services related to gene editing predictions and off-target detection, including:
Take the Next Step: Explore Related Services
In this study, the authors introduced an innovative encoding scheme that transforms each sgRNA-target DNA pair into binary matrices, which serve as the input data for CRISPR-Net. As a classifier, CRISPR-Net applies a binary cross-entropy loss function to the output neurons with sigmoid activation, generating off-target scores for both the target and off-target sequences. In its function as a regressor, the model employs the mean square error (MSE) loss function on the output neurons to compute off-target scores.
CRISPR-Net presents a significant advancement in the field of off-target prediction for CRISPR/Cas9 gene editing. Its superior performance on benchmark datasets and the development of an innovative encoding scheme mark a substantial contribution to improving the precision and safety of CRISPR-based genetic interventions.
The authors have provided a specialized command-line tool designed to quantify off-target effects induced by single-guide RNA (sgRNA). This tool, named CRISPR_Net.py, is specifically designed for the accurate prediction of off-target scores for sequences containing insertions or deletions (Indels) and mismatches. The tool can be accessed via the following link:
https://codeocean.com/capsule/9553651/tree/v3.
The following are the detailed steps for using CRISPR-Net:
Prerequisites:
Before running CRISPR-Net, ensure that Python is installed on the computer (version 3.6 or higher is recommended). Additionally, the required libraries, including scipy, numpy, pandas, scikit-learn, TensorFlow, and Keras, must be installed.
Preparing the Input File:
Prepare an input file named input.csv. In this file, the first column should contain the on-target sequences (i.e., sequences where precise editing is desired), while the second column should list the off-target sequences (i.e., sequences that may be affected non-specifically). Ensure that the data are correctly formatted to allow the tool to process and analyze the sequences efficiently.
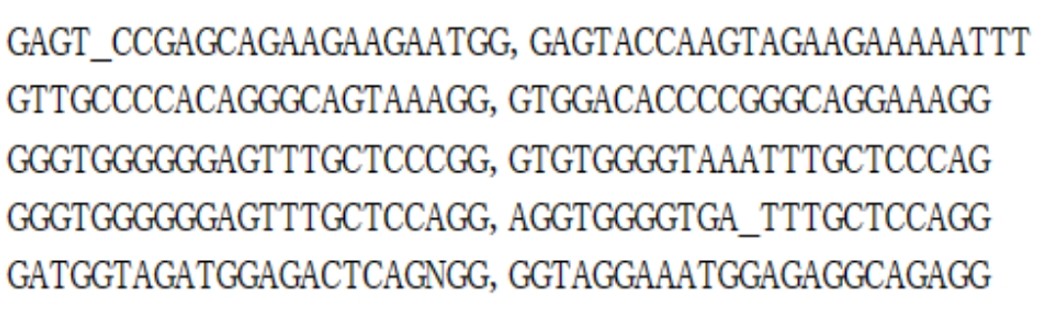
Running CRISPR-Net:
Execute CRISPR-Net by entering the following command in the terminal:
$> python ./CRISPR_net.py input.csv
After execution, the tool will output a file, with the third column displaying the predicted off-target scores generated by the model.

CRISPR-Net provides a robust method for performing genome-wide off-target scoring in advance, allowing researchers to mitigate risks and ensure the safety and efficacy of CRISPR system design and application. After selecting suitable sgRNAs based on predictions, it remains critical to rigorously validate their off-target effects using scientific methodologies.
As a leader in the field of gene editing technology services, CD Genomics is dedicated to providing comprehensive and high-quality technical support to researchers. We offer a wide range of services, including in vivo off-target detection, in vitro off-target detection, and amplicon-based sequencing for editing efficiency evaluation.
References

CD Genomics is transforming biomedical potential into precision insights through seamless sequencing and advanced bioinformatics.
We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy